|
You can do the following manually in Quest by copying the *.QUE Quest file
generated by Platon to a machine with Quest on it and run quest and import the file.
quest -j structure_name < structure_name.que
QUEST Query file
SAVE 3
T1 *CONNSER
AT1 S 2
AT2 N 2
AT3 N 2
AT4 N 2
AT5 N 2
AT6 N 2
AT7 N 1
AT8 C 3
AT9 C 2
AT10 C 2
AT11 C 3
AT12 C 2
AT13 C 3
AT14 C 2
AT15 C 1
AT16 C 1
AT17 C 2
BO 1 10 99
BO 1 12 99
BO 2 11 99
BO 2 16 99
BO 3 8 99
BO 3 17 99
BO 4 11 99
BO 4 14 99
BO 5 13 99
BO 5 17 99
BO 6 9 99
BO 6 11 99
BO 7 9 99
BO 8 13 99
BO 8 15 99
BO 10 14 99
BO 12 13 99
END
QUESTION T1
The Resulting Journal File
COMM +----------------------------------------------------------------------+
COMM | These are comments in the QUEST initialisation file. This file can |
COMM | contain QUEST commands, such as terminal type, that are always read. |
COMM | For more information enter "HELP INITIALISATION FILES" within QUEST. |
COMM +----------------------------------------------------------------------+
COMM | For more information on... |
COMM | the database of CSDS citations, type "HELP DBUSE" |
COMM | starting the graphical interface, type "HELP GRAPHICS" |
COMM | the distributed release notes, type "HELP RELEASE NOTES" |
COMM | the PreQuest data input program, type "HELP PREQUEST" |
COMM | the CIF/MIF output file, type "HELP SAVE" |
COMM +----------------------------------------------------------------------+
COMM | Visit the CCDC web site at: http://www.ccdc.cam.ac.uk/ |
COMM +----------------------------------------------------------------------+
COMM Set better PRINT style:
PRINT 10
COMM +----------------------------------------------------------------------+
SAVE 3
T1 *CONNSER
AT1 S 2
AT2 N 2
AT3 N 2
AT4 N 2
AT5 N 2
AT6 N 2
AT7 N 1
AT8 C 3
AT9 C 2
AT10 C 2
AT11 C 3
AT12 C 2
AT13 C 3
AT14 C 2
AT15 C 1
AT16 C 1
AT17 C 2
BO 1 10 99
BO 1 12 99
BO 2 11 99
BO 2 16 99
BO 3 8 99
BO 3 17 99
BO 4 11 99
BO 4 14 99
BO 5 13 99
BO 5 17 99
BO 6 9 99
BO 6 11 99
BO 7 9 99
BO 8 13 99
BO 8 15 99
BO 10 14 99
BO 12 13 99
END
QUESTION T1
---------+---------+---------+---------+---------+---------+---------+---------+
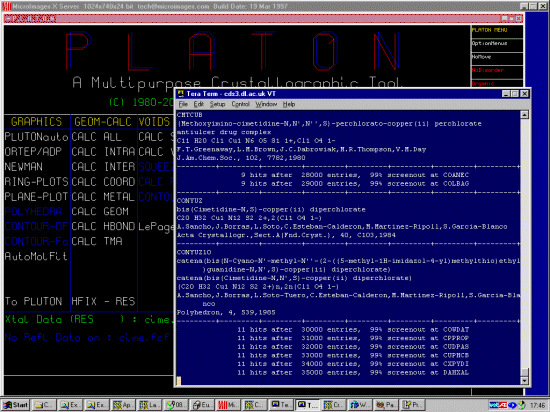
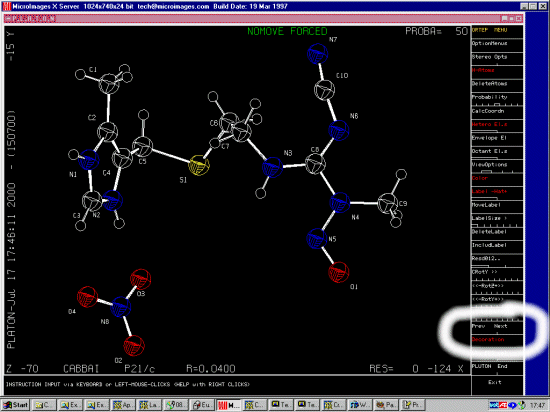
CABBAI
4-(2-(2-Cyano-3-methyl-3-nitroso-1-guanidino)-ethylthiomethyl)-5-methylimidazoli
um nitrate
N-Nitroso-cimetidine nitrate
higher-melting polymorph, potential carcinogenic agent
C10 H16 N7 O1 S1 1+,N1 O3 1-
K.Burns,K.Prout,G.J.Durant
J.Chem.Soc.,Perkin Trans.2, , 1239,1983
---------+---------+---------+---------+---------+---------+---------+---------+
CADVIM
N-Cyano-N'-methyl-N''-(2-(((5-methyl-1H-imidazol-4-yl)-methyl)thio)ethyl)-guanid
ine monohydrochloride monohydrate
Cimetidine monohydrochloride monohydrate
histamine H2-receptor antagonist activity
C10 H17 N6 S1 1+,Cl1 1-,H2 O1
M.Shibata,M.Kagawa,K.Morisaka,T.Ishida,M.Inoue
Acta Crystallogr.,Sect.C (Cr.Str.Comm.), 39, 1255,1983
---------+---------+---------+---------+---------+---------+---------+---------+
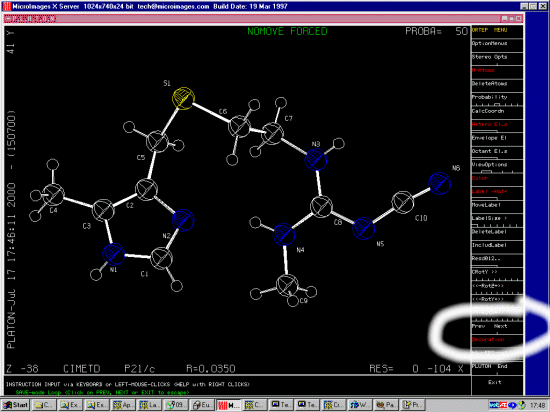
CIMETD
N''-Cyano-N-methyl-N'-(2-((5-methyl-1H-imidazol-4-yl)-methylthio)-ethyl)-guanidi
ne
Cimetidine
monoclinic form A, histamine H2-receptor antagonist activity
C10 H16 N6 S1
E.Hadicke,F.Frickel,A.Franke
Chem.Ber., 111, 3222,1978
---------+---------+---------+---------+---------+---------+---------+---------+
CIMETD01
N-Cyano-N'-methyl-N''-(2-((5-methyl-1H-imidazol-4-yl)methylthio)ethyl)-guanidine
Cimetidine
monoclinic form z, histamine H2-receptor antagonist activity
C10 H16 N6 S1
L.Parkanyi,A.Kalman,B.Hegedus,K.Harsanyi,J.Kreidl
Acta Crystallogr.,Sect.C (Cr.Str.Comm.), 40, 676,1984
---------+---------+---------+---------+---------+---------+---------+---------+
CIMETD02
Cimetidine
N''-Cyano-N'-methyl-N-(2-((5-methyl-1H-imidazol-4-yl)methylthio)ethyl)guanidine
powerful histamine antagonistic activity
C10 H16 N6 S1
S.R.Critchley
Private Communication, , ,1979
---------+---------+---------+---------+---------+---------+---------+---------+
CIMETD03
Cimetidine
N''-Cyano-N'-methyl-N-(2-((5-methyl-1H-imidazol-4-yl)methylthio)ethyl)guanidine
powerful histamine antagonistic activity
C10 H16 N6 S1
R.J.Cernik,A.K.Cheetham,C.K.Prout,D.J.Watkin,A.P.Wilkinson,B.T.M.Willis
J.Appl.Crystallogr., 24, 222,1991
---------+---------+---------+---------+---------+---------+---------+---------+
CIMGUA
2-Cyano-1-methyl-3-(2-((5-methyl-1H-imidazol-4-yl)-methylthio)-ethyl)-guanidine
monohydrate
C10 H16 N6 S1,H2 O1
B.Kojic-Prodic,Z.Ruzic-Toros,N.Bresciani-Pahor,L.Randaccio
Acta Crystallogr.,Sect.B, 36, 1223,1980
---------+---------+---------+---------+---------+---------+---------+---------+
CMTCUA
bis(mu!2$-Cimetidine)-copper(ii) diperchlorate monohydrate
antiulcer drug complex
(C20 H32 Cu1 N12 S2 2+)n,2n(Cl1 O4 1-),n(H2 O1)
F.T.Greenaway,L.M.Brown,J.C.Dabrowiak,M.R.Thompson,V.M.Day
J.Am.Chem.Soc., 102, 7782,1980
---------+---------+---------+---------+---------+---------+---------+---------+
CMTCUB
(Methoxyimino-cimetidine-N,N',N'',S)-perchlorato-copper(ii) perchlorate
antiulcer drug complex
C11 H20 Cl1 Cu1 N6 O5 S1 1+,Cl1 O4 1-
F.T.Greenaway,L.M.Brown,J.C.Dabrowiak,M.R.Thompson,V.M.Day
J.Am.Chem.Soc., 102, 7782,1980
---------+---------+---------+---------+---------+---------+---------+---------+
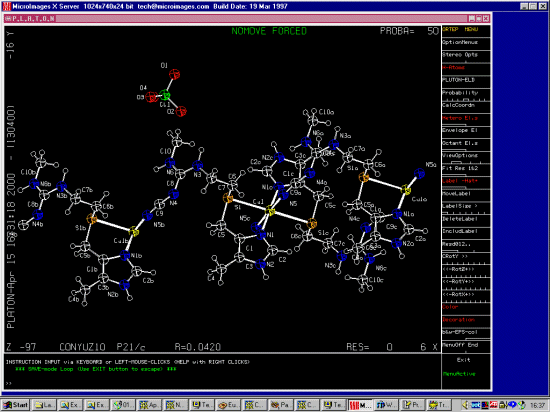
CONYUZ
bis(Cimetidine-N,S)-copper(ii) diperchlorate
C20 H32 Cu1 N12 S2 2+,2(Cl1 O4 1-)
A.Sancho,J.Borras,L.Soto,C.Esteban-Calderon,M.Martinez-Ripoll,S.Garcia-Blanco
Acta Crystallogr.,Sect.A (Fnd.Cryst.), 40, C103,1984
---------+---------+---------+---------+---------+---------+---------+---------+
CONYUZ10
catena(bis(N-Cyano-N'-methyl-N''-(2-((5-methyl-1H-imidazol-4-yl)methylthio)ethyl
)guanidine-N,N',S)-copper(ii) diperchlorate)
catena(bis(Cimetidine-N,N',S)-copper(ii) diperchlorate)
(C20 H32 Cu1 N12 S2 2+)n,2n(Cl1 O4 1-)
A.Sancho,J.Borras,L.Soto-Tuero,C.Esteban-Calderon,M.Martinez-Ripoll,S.Garcia-Bla
nco
Polyhedron, 4, 539,1985
---------+---------+---------+---------+---------+---------+---------+---------+
DEFWEQ
bis(N-Cyano-N'-methyl-N''-(2-(((5-methyl-1H-imidazol-4-yl)-methyl)-thio)-ethyl)-
guanidine)-copper(ii) dinitrate
bis(Cimetidine)-copper(ii) dinitrate
(C20 H32 Cu1 N12 S2 2+)n,2n(N1 O3 1-)
L.Soto,J.Borras,A.Sancho,A.Fuertes,C.Miravitlles
Acta Crystallogr.,Sect.C (Cr.Str.Comm.), 41, 1431,1985
---------+---------+---------+---------+---------+---------+---------+---------+
DOKGAL
bis(Cimetidine)-cobalt(ii) diperchlorate
bis(N-Cyano-N'-methyl-N''-(2-((5-methyl-1H-imidazol-4-yl)-methylthio)ethyl)guani
dine)-cobalt(ii) diperchlorate
(C20 H32 Co1 N12 S2 2+)n,2(Cl1 O4 1-)
A.Sancho,L.Soto-Tuero,A.Cantarero,M.I.Arriortua,J.M.Amigo
Z.Kristallogr., 173, 155,1985
---------+---------+---------+---------+---------+---------+---------+---------+
DOKGEP
bis(Cimetidine)-nickel(ii) diperchlorate
bis(N-Cyano-N'-methyl-N''-(2-((5-methyl-1H-imidazol-4-yl)-methylthio)ethyl)guani
dine)-nickel(ii) diperchlorate
(C20 H32 N12 Ni1 S2 2+)n,2(Cl1 O4 1-)
A.Sancho,L.Soto-Tuero,A.Cantarero,M.I.Arriortua,J.M.Amigo
Z.Kristallogr., 173, 155,1985
---------+---------+---------+---------+---------+---------+---------+---------+
GAXVUW
catena(bis(N-Cyano-N'-methyl-N''-(2-((5-methyl-1H-imidazol-4-yl)methylthio)ethyl
)guanidine-N''',N'''',S)-nickel bis(tetrafluoroborate))
(C20 H32 N12 Ni1 S2 2+)n,2n(B1 F4 1-)
J.M.Amigo,M.M.Reventos,A.Sancho,L.Soto-Tuero,A.Cantarero
Z.Kristallogr., 180, 123,1987
---------+---------+---------+---------+---------+---------+---------+---------+
GEWYUC
catena(bis(mu!2$-Cimetidine-N,N',S)-copper(ii) sulfate nonahydrate)
catena(bis(mu!2$-N-Cyano-N'-methyl-N''-(2-((5--methyl-1H-imidazol-4-yl)methylthi
o)ethyl)guanidine-N,N',S)-copper(ii) sulfate nonahydrate)
(C20 H32 Cu1 N12 S2 2+)n,n(O4 S1 2-),9n(H2 O1)
L.Soto,J.P.Legros,A.Sancho
Polyhedron, 7, 307,1988
---------+---------+---------+---------+---------+---------+---------+---------+
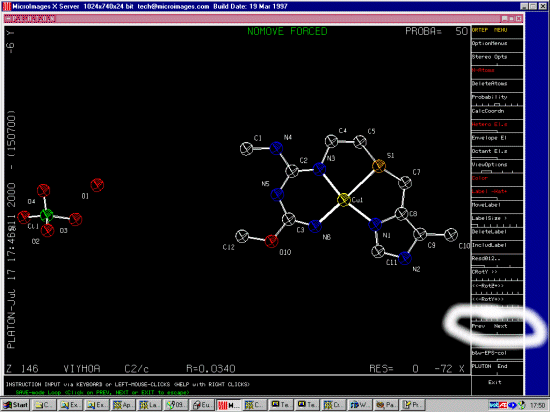
VIYHOA
(5-Methylcimetidine-S,N,N',N'')-copper(ii) perchlorate hemihydrate
C11 H19 Cu1 N6 O1 S1 1+,Cl1 O4 1-,0.5(H2 O1)
A.M.Bianucci,F.Demartin,M.Manassero,N.Masciocchi,M.L.Ganadu,L.Naldini,A.Panzanel
li
Inorg.Chim.Acta, 182, 197,1991
---------+---------+---------+---------+---------+---------+---------+---------+
VIYHUG
(5-Methylcimetidine-S,N,N',N'')-copper(ii) iodide hemihydrate
C11 H19 Cu1 N6 O1 S1 1+,I1 1-,0.5(H2 O1)
A.M.Bianucci,F.Demartin,M.Manassero,N.Masciocchi,M.L.Ganadu,L.Naldini,A.Panzanel
li
Inorg.Chim.Acta, 182, 197,1991
|