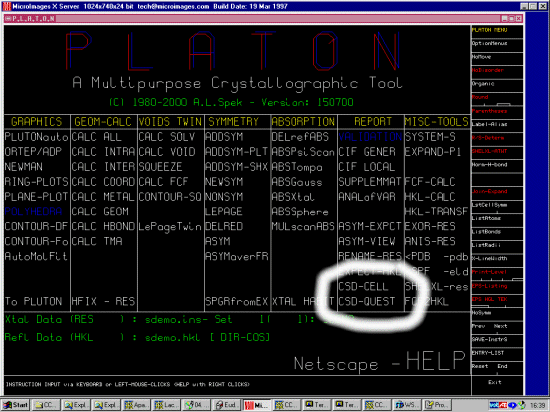
Another mega-powerful time saving feature of platon is the ability to
act as a user-friendly interface for the Cambridge Database Quest program to
search on related organic and organometallic phases. Which thus offers the
ability to easily determine if the structure has already been solved via
giving it the cell, or "your" solved structure.
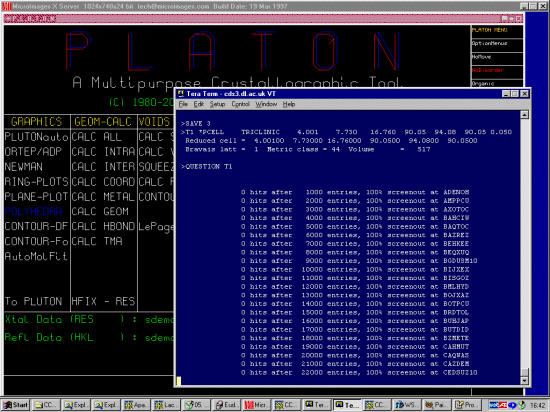
In the following case, we will give Platon the cell information of an
organic phase to see if the structure may have already been solved.
If you are already running platon on a UNIX computer where the CSD and Quest
already been installed, this is extremely trivial.
Note: If searching on a cell obtained from Powder diffraction data, and you do not
get any hits. It is advisable to edit the *.QUE file and change the tolerance from
0.01 to 0.05; then manually run Quest. This is described in more detail at the bottom of this
tutorial.
Here we will deal with the case where the user is on a Windows PC, and the CSD database is
on a remote UNIX machine where Platon has been compiled. (it is possible to
generate the Quest query on Platon for Windows and ftp it over to run
manually in quest - using a command line similar to quest -j structure_name < structure_name.que)
NOTE: With quest, make sure it is not automatically going into Xterm/Menu mode or the
following will not work. By default, Quest should not go into Xterm/Menu mode but the
administrator may have set it up like this to save on a few keystrokes.
If you have not done already, information on settup up for Secure FTP and Secure X sessions
from a Windows machine, refer:
If a UK based academic or student, you can obtain free and easy
access to the Cambridge and other structure databases via the EPSRC funded
CDS - Chemical Database Service
(free registration is done On-line)