CCP14
Tutorials and Examples
Peak Fitting using Xfit-Koalariet (Coelho and Cheary) for Win95/NT
Fundamental Parameters Peak Profiling on lab Powder XRD data for Ab-initio Indexing and Structure Solution
The CCP14 Homepage is at http://www.ccp14.ac.uk
[The reference to use for XFIT or FOURYA in any resulting publications is:
Cheary, R. W. & Coelho, A. A. (1996). Programs XFIT and FOURYA, deposited in CCP14 Powder Diffraction Library,
Engineering and Physical Sciences Research Council, Daresbury Laboratory, Warrington, England.
(http://www.ccp14.ac.uk/tutorial/xfit-95/xfit.htm)]
The Problem
Following is a real world example of using Fundamental Parameters peak profiling and XFIT to obtain
peak information for auto-indexing and structure factor extraction for (in this case) Patterson based
structure solution. Peaks are defined visually. If there is a significant misfit, this could
imply an extra peak is present but care needs to be taken. For difficult problems, indexing
in done iteratively with the peak fitting to check for possible impurity phases.
The data is of a new Lithium Titanate (I.E. Grey, L. M. D. Cranswick, C. Li,
L. A. Bursill, and J. L. Peng, "New Phases Formed in the Li-Ti-O System under
Reducing Conditions", Journal of Solid State Chemistry, 138, 74-86 (1998))
The advantage of fundamental parameters over Empirical peak fitting is more
confidence in fitting real peaks due to being inherantly able to model real
peak asymmetry due to the diffractometer geometry; and more of a handle in terms of trying
to understand what any misfits could be due to. Also, as you are trying to refine on
physically meaningful parameters, parameters such as crystallite size and crystallite
strain can be used as to describe the width/shape of all the peaks resulting in only
2 parameters per peak instead of 4 or greater with empirical peak modelling methods.
Download Data
Click here to download a zip file of the Liti data
The powder X-ray Diffraction data is a fast 20 minute continuous scan from 5 to 70 degrees run
on a Philips Diffractometer. The reason this data was used was that the sample degraded somewhat
by the time a slow Rietveld quality step scan had been run. Rather than twiddle
thumbs for the night, it was decided to use the continous scan data. It eventuated
that not only could this quick continuous scan data index the phase and assign a space group
from the absences; but fitted intensities allowed the structure to be solved with Shelx using
Pattersons down to the Oxygens. A new preparation, Rietveld quality XRD data, combined
with difference map Fourier contour maps inside the CSRIET software, allowed refinement
down to the Lithiums. Neutron data was used to confirm the position and reliability of
the Lithium positions.
While the Philips APD 3.6 for DOS software was used for peak profiling on the original
structure solution, this goes through the methodology using freely available software.
The scan was collected on a Philips 1710 system with 1050 goniometer
(173mm goniometer radius) with Cu LFF X-ray tube at 40mA and 40kV,
incident beam Sollers slit with 5.1 degree acceptance angle, no
diffracted beam Sollers
slit (equivalent of 9.1 degree acceptance angle)
1 degree fixed divergence slit, 0.45mm receiving slit, 1 degree
scatter slit, diffracted beam curved graphite monochromator and
proportional counter. Continous 20 minute scan from 5 to 70 degrees
at 0.025 steps. Slit lengths are 12mm (wide optics) and sample
length is ~25mm.
One thing to keep in mind is that some of the misfit could be slight
degradation of the sample. Which can make things awkward. Please note
that if there is a "new" effect that is not included in the fundamental
parameters
Note: Remember that when you add a peak using the fundamental parameters
method and XFIT, you have to go back into "File Details, Peaks, Fundamental
Parameters Peaks"; change the strain from the default of 0, to match the
peaks you are linking this to (or 0.01 if freely refined); change the crystallite size to the
peaks you are linking this to; "Use" the peak, then enable the refinement flags
for these newly inserted peaks. Tedious compared to the Empirical peak fitting
method but such is life.
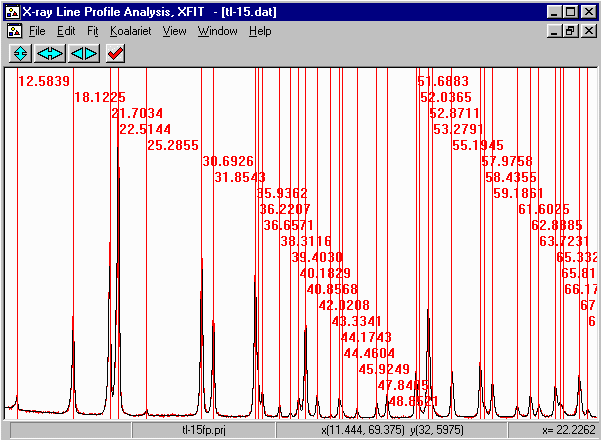
- Run XFIT, (maximise the screen if you wish)
and do File, Load Data and open
the tl-15.dat dataset. You can maximize the data to
fit the entire XFIT screen. It should look something like
the following. (if you can't see the active mouse output on
the bottom right of the XFIT screen - maximise your XFIT
window)
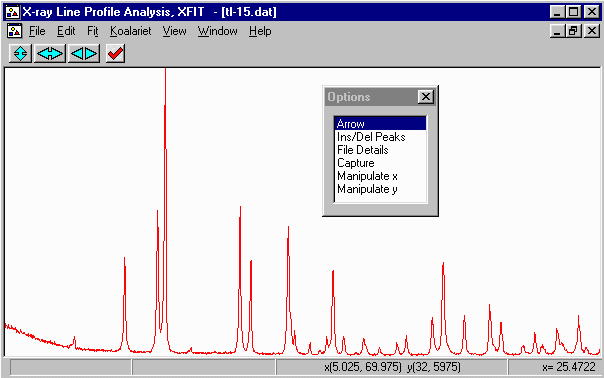
- Examining the data, with fundamental parameters fitting, you should be able to
fit the entire pattern. Also, it can be quite tedious zooming and fitting small
sub-sections later if the peaks area linked by the same crystallite size and
strain code-words. This is because XFIT only refines on peaks visible on the
screen; but if linked to peaks outside the screen, things go a bit haywire.
Thus if you do zoom up to refine a subset of peaks, you have to make sure any linked
parameters are only on the peaks visiible on the screen that you are refining.
Setting up the Cu X-ray Emission Profile, Diffractometer and Background Parameters
- As the characteristic X-rays from the Copper X-ray tube consist
of alpha1 and alpha2 wavelengths, the wavelength being used has to be
assigned so that the peaks will be profiled correctly.
From the Options box, select File Details elect
the Assign LAMs to Files and borwse to c:\koalarie directory
and select the cuka_2.lam file.
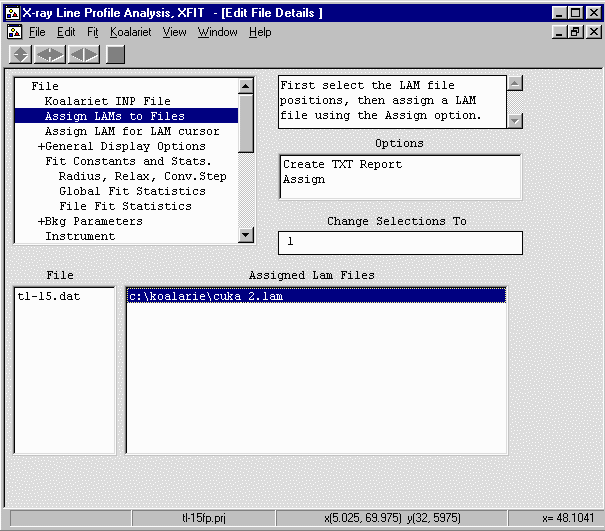
- Go to the Fit Constants and Stats; Radius, Relax Conv.Step
and input the goniometer radius of 173mm (highlight the Radius
value, type 173 in the "Change Selections To" box and press the ENTER/RETURN
key).
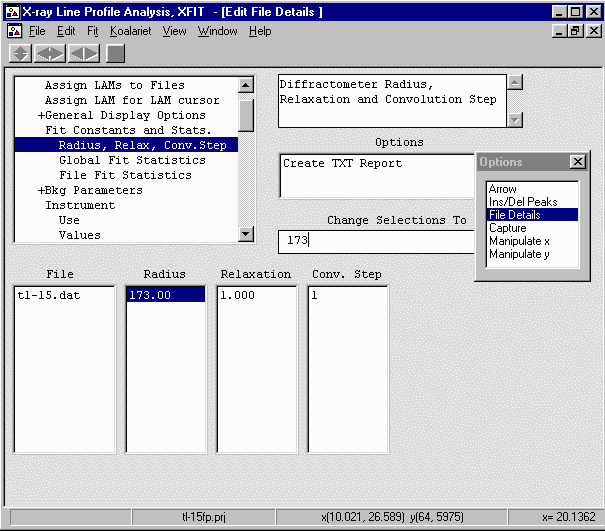
- Continue onto Bkg Parameters and Set the Polynomial order to 5 so it can
handle the low angle part of the background.
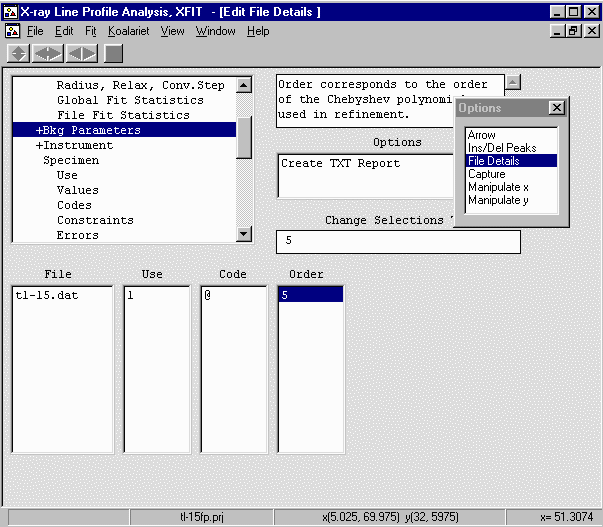
- Continue onto Instrument, Values and set the SW (receiving slit) to 0.45mm,
FSFA (Divergence slit) to 1 degree, and X-ray Tube Target (long fine focus) to 0.04mm.
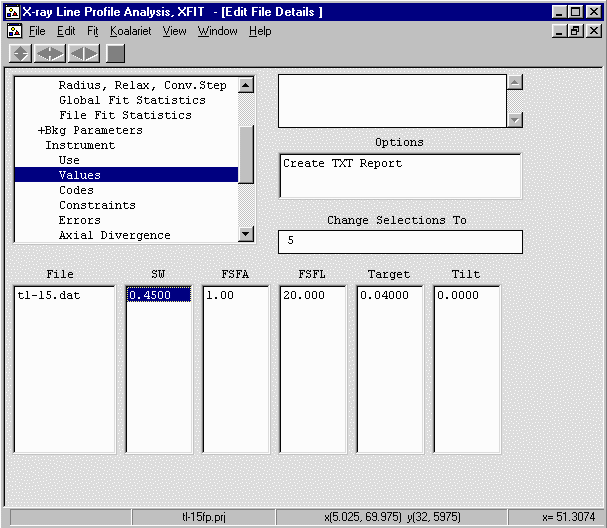
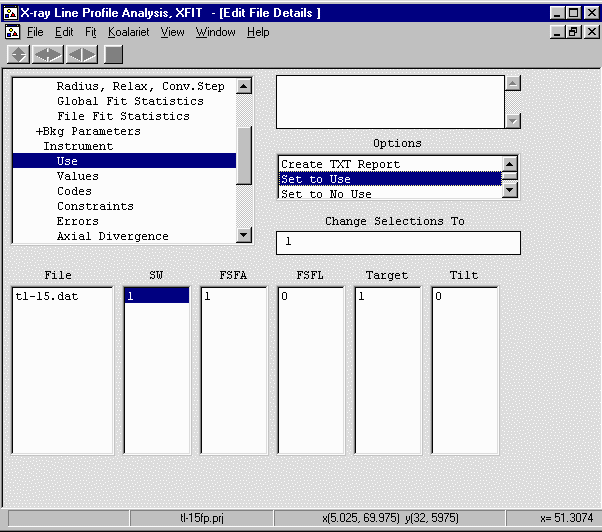
- Go up one to Instrument, Use and enable the above parameters (SW, FSFA, Target)
by seleting each 0 value, then Set to Unique Codes from the "Change Selections To" box to
convert it to a "1" (Use) value.
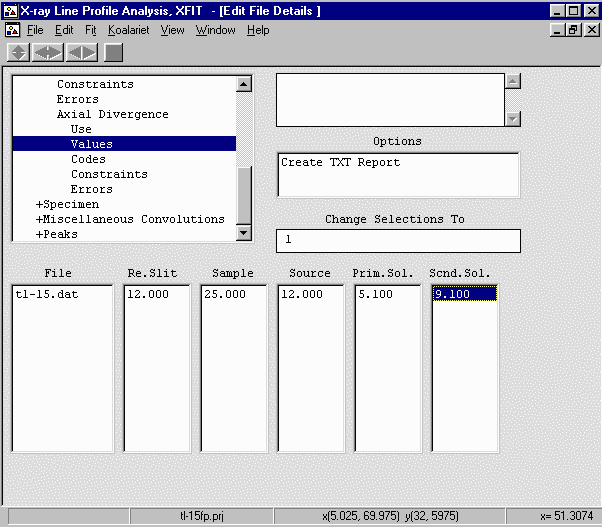
- Go down to Instrument, Axial Divergence, Values and set the Re.Slit (length) to 12mm;
Sample (Length) to 25mm; Source (length) to 12mm; Prim.Sol. (Incident Beam Sollers Slit) to 5.1
degree acceptance angle; Scnd.Sol. (Diffracted Beam Sollers Slit) to 9.1 degree acceptance angle
which is the same as saying there is no Diffracted Beam Sollers Slit.
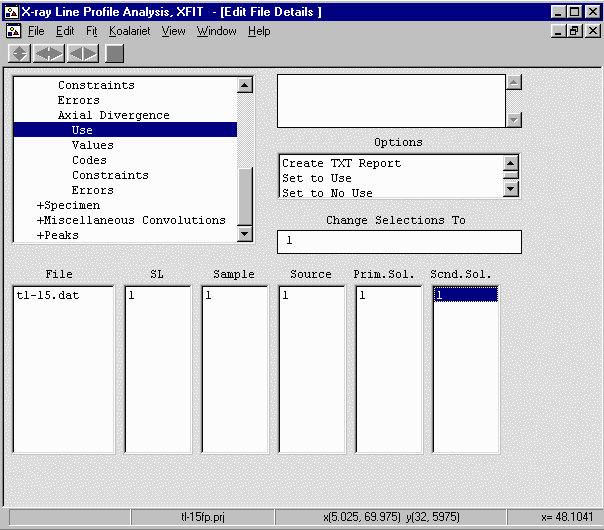
- Go up one to Instrument, Use and enable the above parameters (Re.Slit; Sample; Source;
Prim.Sol.; Scnd.Sol.) by seleting each 0 value, then Set to Unique Codes from
the "Change Selections To" box to convert it to a "1" (Use) value.
- Congrats! You have setup the machine and background options for this sample. Now it is
a matter of inserting, configuring and refining the peaks.
Selecting and Fitting the Peaks
- In the Options box, select arrow use
the left mouse key to zoom to 11 degrees and the right mouse button to zoom to 26
degrees. Press s if you miss zoom to get back to the full screen.
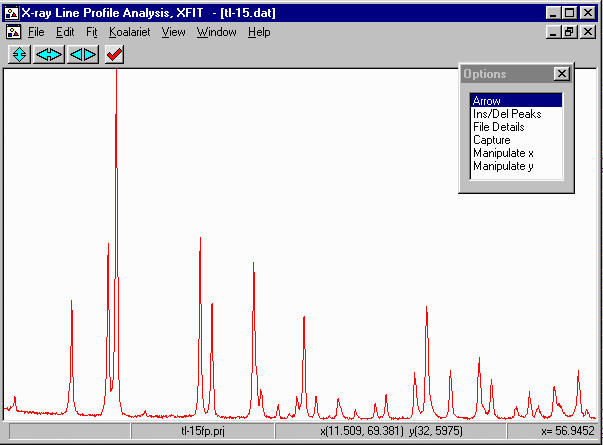
- Select Option, Ins/Del Peaks, Fundamental Parameters Peaks, and insert peaks
by clicking the left mouse button.
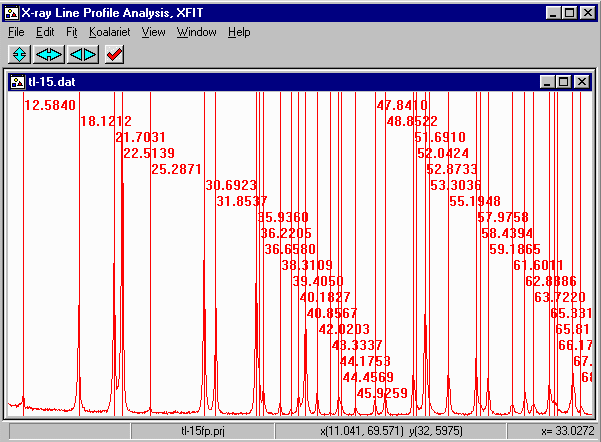
- From the Options box, select File Details, Peaks, Values. Select
all the strain value (click and drag) and give the strain values a value greater
than zero (i.e., 0.01)
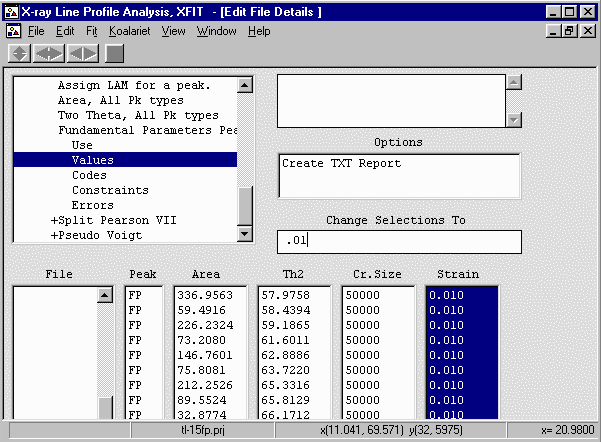
- Go up one menu option to Use and select the Cr Size and Strain values,
Set to Use from the "Change Selections To" box to convert it to a "1" (Use) value.
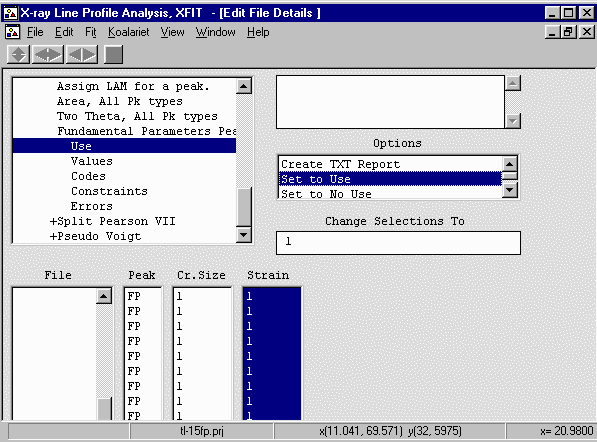
- Go down two menu options to Codes and select the Cr Size and Strain columns,
Set the codewords cry and strain in the "Change Selections To"
box to convert links the size and strain for all peaks to be identical. The assumption
here is that the peaks share identical crystallites size/strain values which may not
be the case. The size/strain can be refined individually for each peak. This speeds
up the calculation and can result in more reliable values.
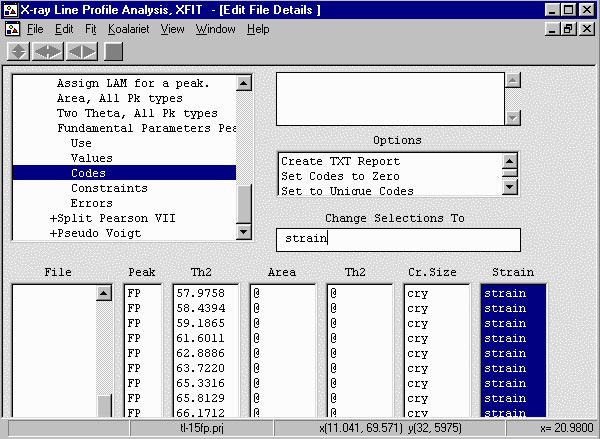
- From the menu select, Fit, Fit Marquardt.
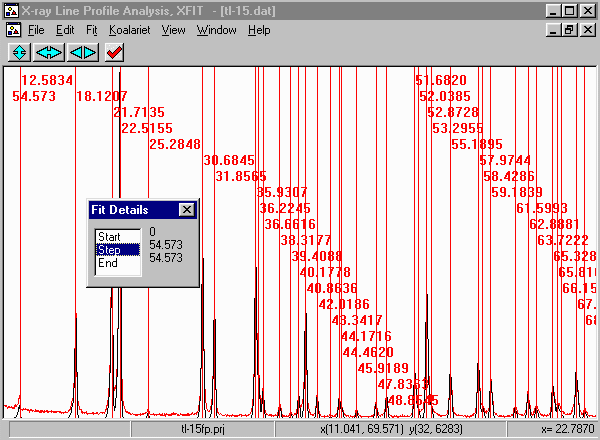
- Then click on start to start fitting and then Yes to keep
the refined parameters.
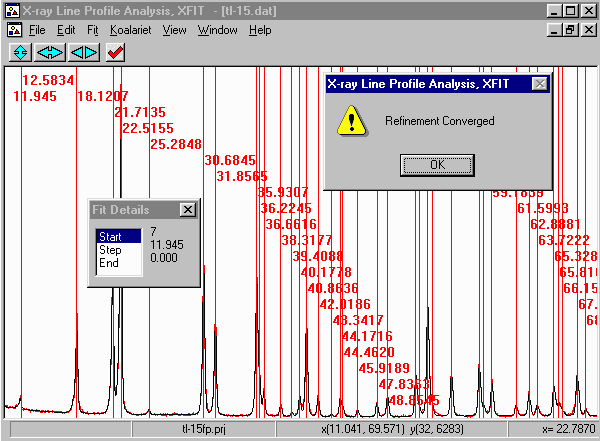
- Note that misfits are due to either problems with the diffractometer system (e.g.
misalignment or packing of the sample) , or with the sample, (e.g. more than one peak
present, disorder, new/novel form of strain or disorder, degradation of the sample).
In this case slow degradation of the sample is the most likely cause of the misfit.
It is possible to possibly correct for this by refining various XRD geometry to unreasonable
values to some extent; but we will not do this here.
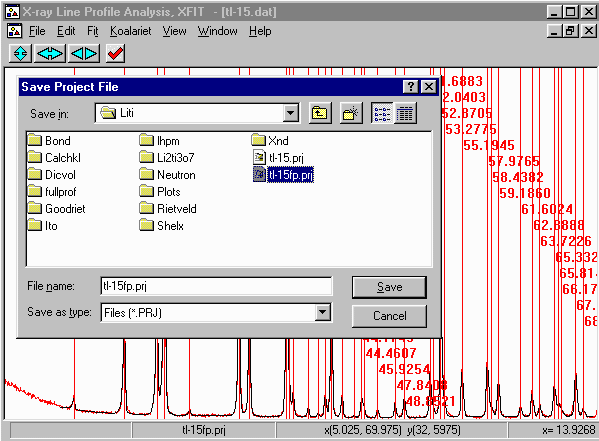
- Just in case XFIT might feel like crashing, save the project file
via the menu using File, Save Project As
- To freely release the crystallite size and strain to refine for each peak,
Options box, select File Details, Peaks, Fundamental Parameters Peaks,
Values. Select the Crystallite Size and Strain codewords (cry, strain) and set them
to using the Set to Unique Codes
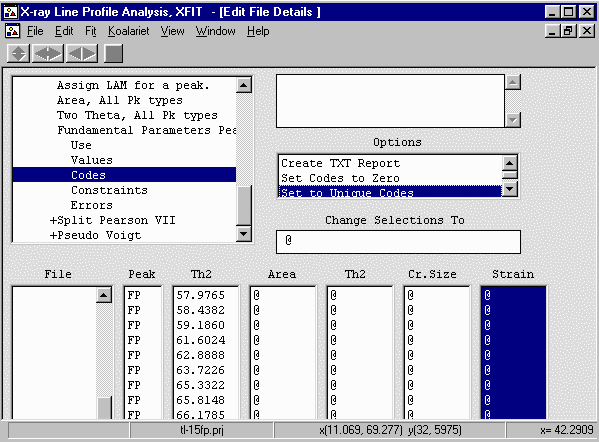
- From the menu select, Fit, Fit Marquardt. Then click
on start to start fitting and then Yes to keep
the refined parameters. The fit only went down from 11.9 to 11.3
which is not that big a change.
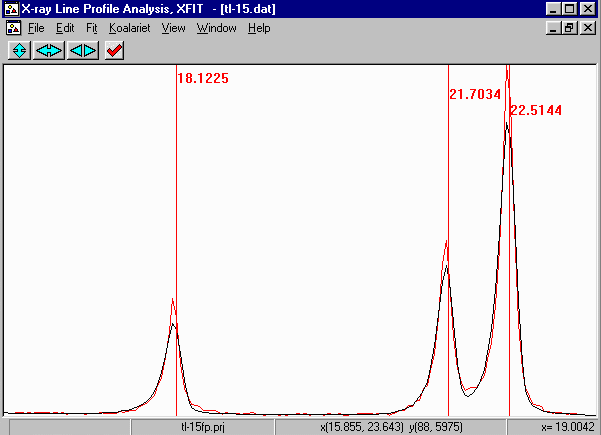
Examining the Peak Fit
- Zooming up allows you to see the goodness of fit visually. you will note
that the fit does not look all that great. This is probably due to degradation of
the sample or some form of strain that XFIT is not modelling. Refining on the
Sollers slit accpetance angles gives values close to the "official" value for
this geometry, implying peak asymmetry is fitting correctly. However, note that
a FP fit of the 21.7 and 22.5 peaks imply that the peak inbetween them might be
just a bit of noise. The mathematical/empirical fit migth make this situation
far more difficult to interpret. Indeed in the
Empirical Fitting Using XFIT tutorial - use of
an Split Pearson peak shape implied there could be a peak between these
two intense peaks.
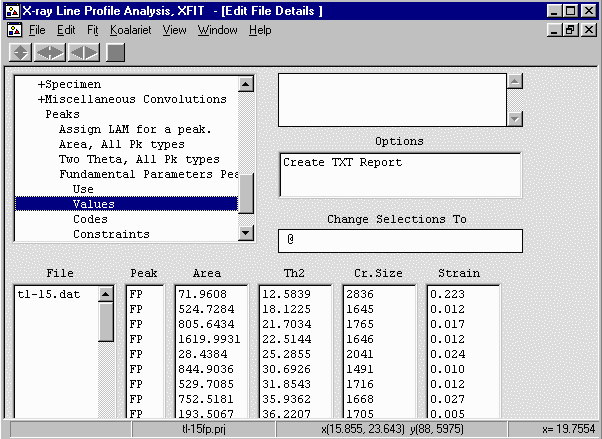
Obtaining Results
- This results in a list of peaks with ESDs. To generate ESDs, re-fit the range of peaks
of interest, then from the menu do Fit, Errors.
Go into File Details, Peaks, Fundamental Parameters Peaks, Values, Create Text Report to get
the results. Comparing the ESDs with that of the empirical peak fitting on the same sample
can be quite revealing; especially the ESDs on FWHM and Shape using Empirical Peak Fitting.
File Peak Area ESD Th2 ESD
tl-15.dat FP 73.7972 7.04 12.5833 0.0152
FP 525.1695 11.33 18.1219 0.0031
FP 808.8842 13.64 21.7033 0.0024
FP 1617.7630 18.58 22.5138 0.0016
FP 29.2490 4.03 25.2852 0.0212
FP 842.7766 13.50 30.6925 0.0022
FP 530.1695 10.90 31.8536 0.0027
FP 755.3265 15.95 35.9364 0.0031
FP 194.3486 11.40 36.2205 0.0091
FP 135.4252 6.50 36.6579 0.0072
FP 61.5260 4.51 38.3112 0.0105
FP 23.0393 3.40 39.4055 0.0230
FP 115.8754 5.67 40.1829 0.0076
FP 557.0820 11.02 40.8568 0.0027
FP 116.2905 5.50 42.0205 0.0069
FP 12.0952 2.91 43.3329 0.0378
FP 106.6417 6.94 44.1745 0.0104
FP 50.8949 6.00 44.4607 0.0192
FP 43.0744 3.88 45.9254 0.0139
FP 81.4082 4.82 47.8408 0.0091
FP 126.3955 5.72 48.8521 0.0068
FP 259.5472 8.78 51.6883 0.0057
FP 46.9653 5.74 52.0403 0.0238
FP 715.0383 13.28 52.8705 0.0030
FP 82.8354 6.46 53.2775 0.0153
FP 278.5762 8.10 55.1945 0.0045
FP 333.2409 9.23 57.9765 0.0045
FP 56.8873 5.20 58.4382 0.0181
FP 222.0052 7.47 59.1860 0.0054
FP 69.8183 4.94 61.6024 0.0125
FP 144.1896 6.40 62.8888 0.0074
FP 74.4343 5.09 63.7226 0.0128
FP 212.7719 7.90 65.3322 0.0068
FP 90.6220 6.80 65.8148 0.0177
FP 33.6637 5.76 66.1785 0.0374
FP 301.1027 8.75 67.6631 0.0049
FP 47.2308 5.10 68.5051 0.0179




















