CCP14
Tutorials and Examples
Peak Fitting using Xfit-Koalariet (Coelho and Cheary) for Win95/NT
Fitting Assymmetrical (Lab XRD) peaks - Y2O3
The CCP14 Homepage is at http://www.ccp14.ac.uk
[The reference to use for XFIT or FOURYA in any resulting publications is:
Cheary, R. W. & Coelho, A. A. (1996). Programs XFIT and FOURYA, deposited in CCP14 Powder Diffraction Library,
Engineering and Physical Sciences Research Council, Daresbury Laboratory, Warrington, England.
(http://www.ccp14.ac.uk/tutorial/xfit-95/xfit.htm)]
Why Bother?
In the following example, we are going to go through
what it takes to fit an assymmetric peak with XFIT.
In this case a low angle lab XRD peak that has low angle
peak assymmetry. This data was collected with a
Philips 1050 goniometer with Cu LFF X-ray tube, 5 degree
incident and diffracted beam Sollers, 1 degree divergence
slit, 0.2mm receiving slit, 1 degree scatter slit,
diffracted beam curved graphite monochromator and proportional
counter.
Download Data
Click here to download a zip file of the Y2O3 data
- Run XFIT, (maximise the screen if you wish)
and do File, Load Data and open
the y2o3.dat data set. You can maximize the data to
fit the entire XFIT screen. It should look something like
the following. (if you can't see the active mouse output on
the bottom right of the XFIT screen - maximise your XFIT
window)
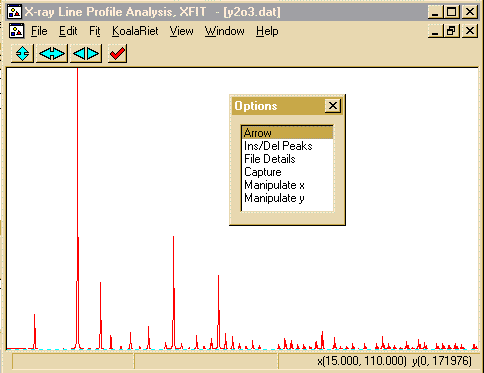
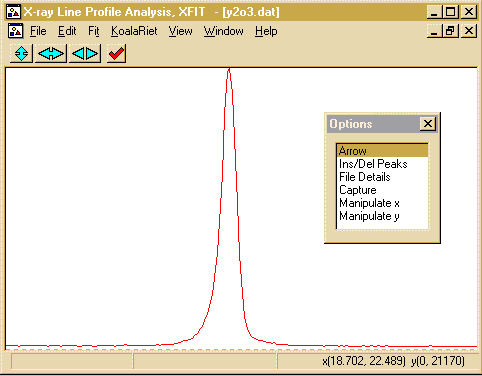
- Zoom into the area around 18 and 22 degrees. This is done
using the left mouse button to define the lower angle and the
right mouse button to define the upper angle.
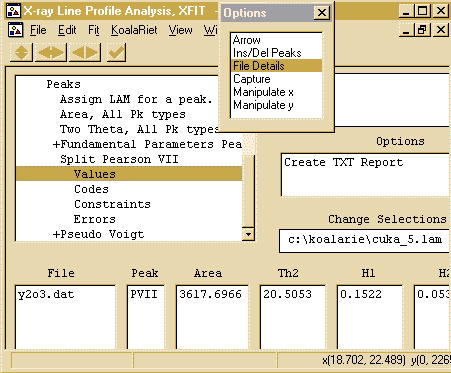
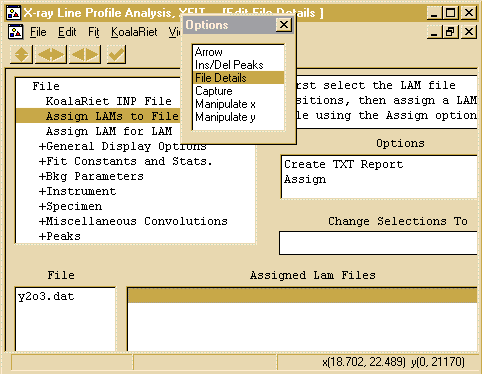
- To tell XFIT that you are using Copper X-ray data from
a lab source, use the Options box to go into the
File Details, and click on the Assign LAMs to
Files
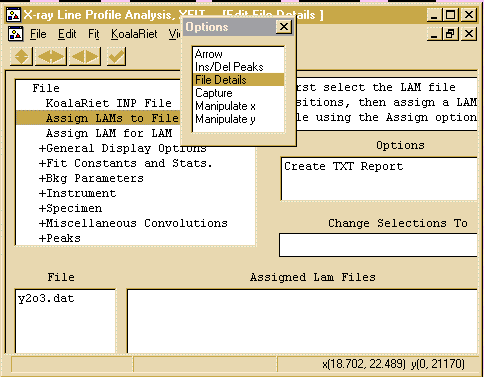
- Then tell click below the Assign LAM Files to the right
of the y2o3.dat file so that you bring up the Assign option
in the Options area.
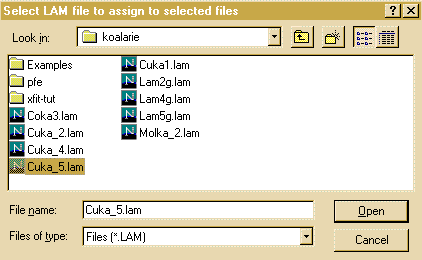
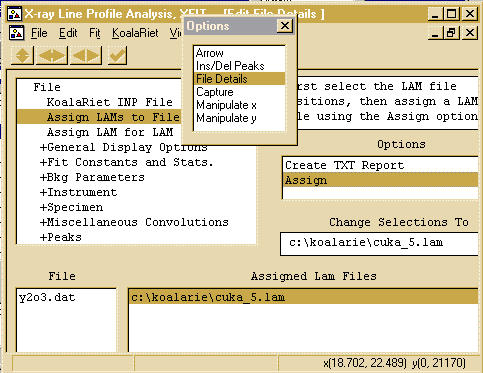
- Then click on the Assign option in the Options area
to bring up the browse box, and go to the c:\koalarie directory
and assign/load the Cuka_5.lam. This describes the
emmission spectrum for the tube (actually 5 characteristic lines -
you can do the traditional approximation of Alpha1 and Alpha2 by
loading the Cuka_2.lam file)
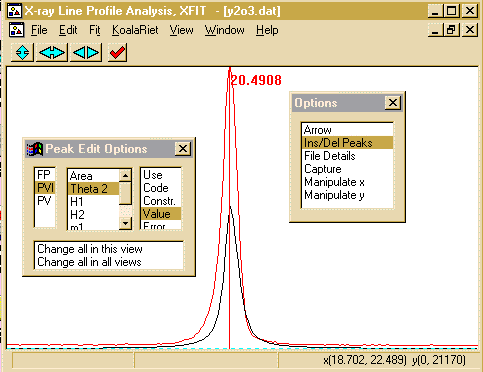
- Go to the Options Box, click on Ins/Del peaks to bring
up the Peak Edit Options box. As peak has low angle
peak assymmetry, select the PVII Split Pearson VII
peak fitting model. Then with use the mouse and left button to
select ONLY the alpha-1 component of the peak (easy here
as they have merged to look like one peak). If you incorrectly
add a peak, put the mouse over the offending peak and click the
right mouse button to delete it.
- From the top menu bar, select Fit, Fit Marqardt.
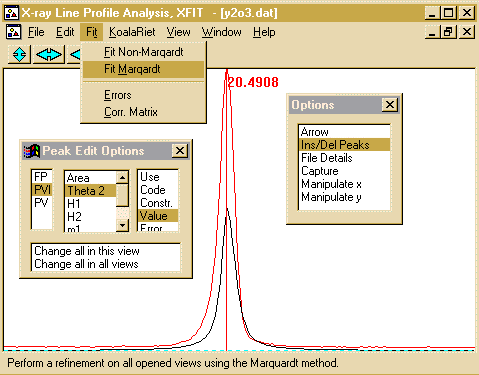
- This brings up Fit Details box. You have the option to
step through the fitting cycles one at a time. You can stop
on the next cycle - and keep all the information refined up to that
point.
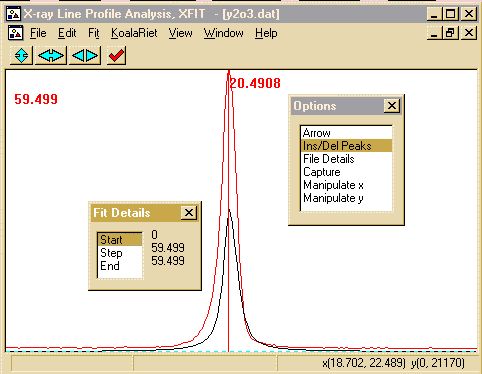
- Click on the Start to start the peak refinement. This
can go rather quickly and when the refinement converges, you will
get a Refinement converged prompt.
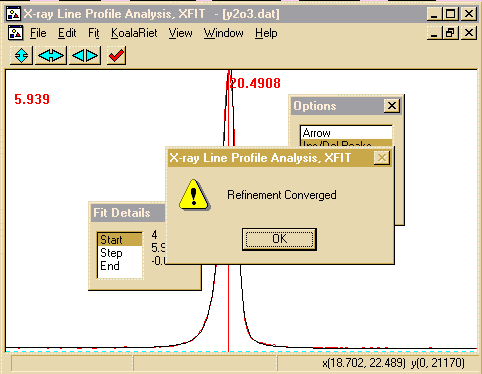
- Press OK and Accept the Refined parameters.
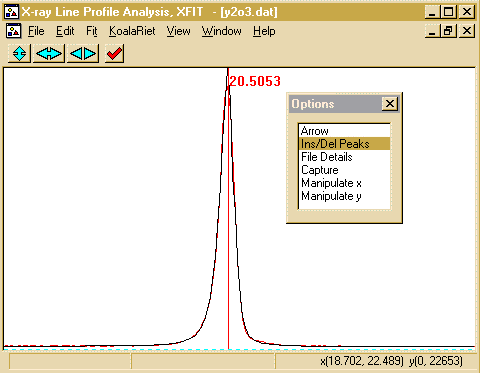
- You can calculate errors by using the Fit, Errors
option. To view the results, which includes the peak Position,
FWHM (Full Width Half Maximum), Shape and Area; go into
File Details, Peaks, Split Pearson VII, Values and the
peak results will be displayed. Clicking on Create TXT
Report will put the peak into an ASCII file that can then be
saved or copied using [CNTRL] C. You can also use File,
Save Project As to save your work for future reference or
to use the fit as a template for other similar files.