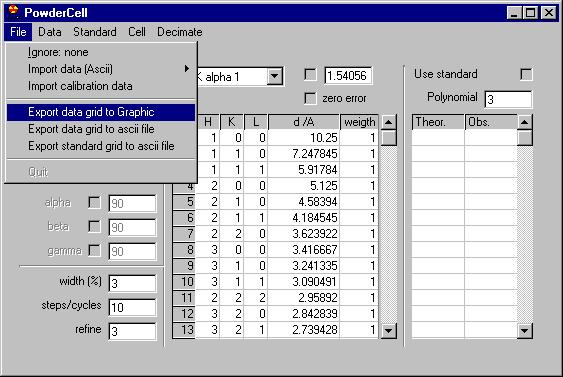
|
 |
 |
|
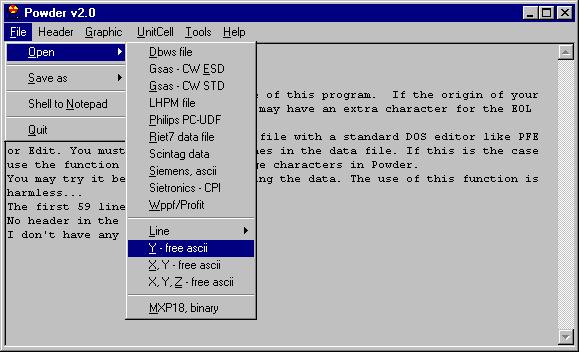
Then open the file as Ascii, and choose the file in question. |
|
|
|
 |
|
|
|
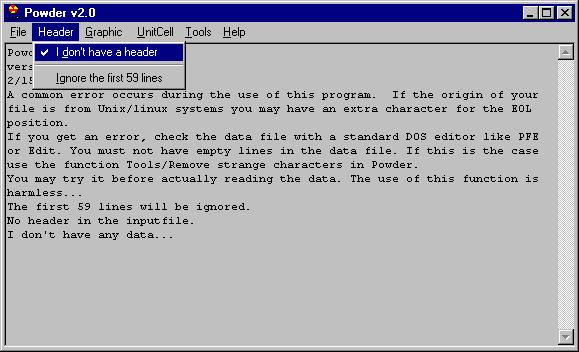
If this is successful you will see some messages
(this is what is called Report Pad in this program), in this case what we
can see is:
-------------------------------------------------
Y data file, ascii
The filename is C:\WINDOWS\Desktop\NAC-test-2.UXD
59 line(s) will be ignored.
The number of points is: 6501
Done...
2/15/99 5:26:39 PM
--
NOTE: I have noticed a frequent error in the use of this program. When
the data file has been e-mailed without being zipped an extra EOL character
appears (I guess it is related to different definition of EOL for DOS and
Windows ?). If you look at this kind of file with DOS/Edit you can see
alternating empty lines with data. These empty lines will "confuse" the program.
Therefore you should remove them by the command Tools/Remove strange characters.
The same procedure should be used for files coming from Unix and flavors
of Unix.
--
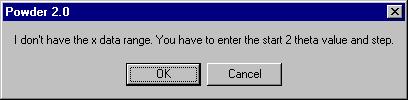
We can directly inspect the data once read by choosing Graphic in the menu.
We get this message |
|
|
|
 |
|
|
|
and we answer 10 for 2 theta and 0.02 for step
(this message comes when the program does not know the starting value and
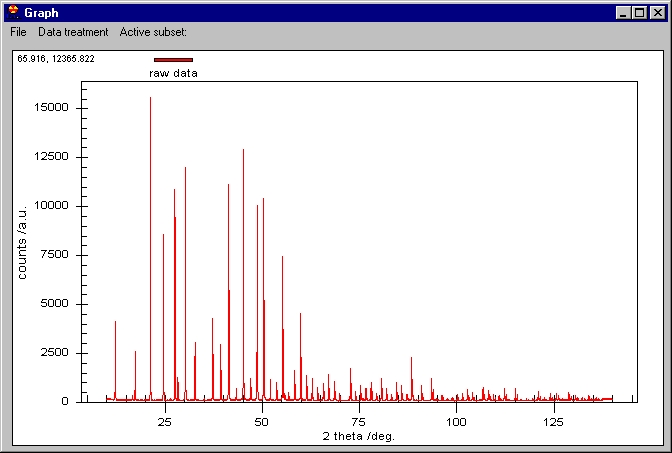
step). Following, the Graphic window is shown: |
|
|
|
 |
|
|
The scale is automatically adjusted to fit all
the data. In the top left corner we can see two values 65.916, and 12365...:
these are the coordinates of the mouse on the screen. You can roughly inspect
the data by this method.
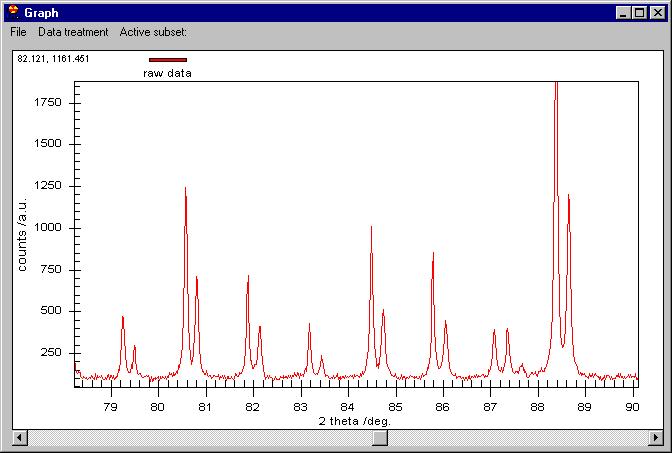
You can zoom as you like by dragging the mouse on the screen: |
|
|
|
 |
|
|
|
On the zoomed graph a horizontal bar appears.
Clicking on it you can move to the right or left by keeping the same zoom
as here. You can go back to the initial zoom by right-clicking on the graph
area and choosingUndo zoom.
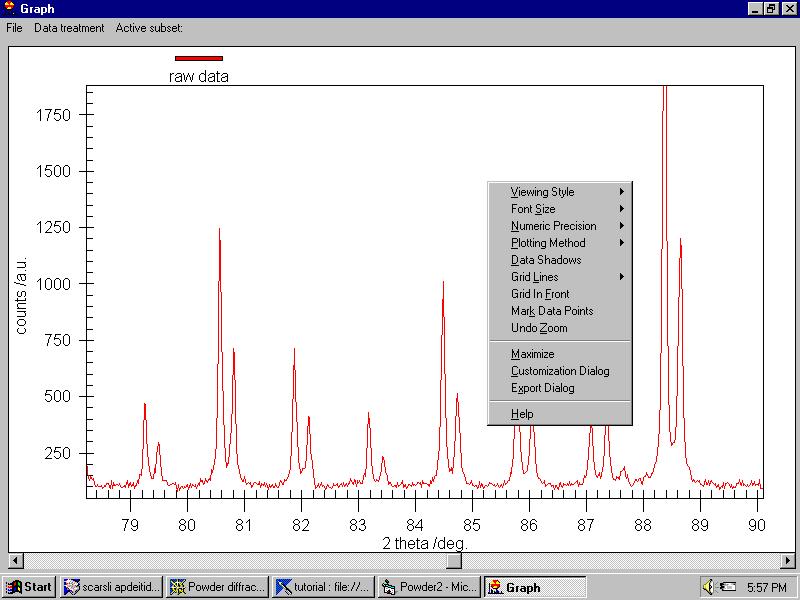
Several other menus are available here, most of them are easy to understand.
When you right-click on the graph an important menu allows changing some
parameters: |
|
|
|
 |
|
|
Note that we can resize the graph at any time.
The menu (which can be invoked -for people hating the mouse- by File/General
Options command as well) contains:
Viewing style (monochrome or color)
Font size
Numeric precision
Plotting method
Grid lines
Grid in front
Mark data points
Undo zoom
Maximize
Customization dialog
Export dialog
Help
Some of these menus are self explanatory; note that Mark Data Points
is extremely slow.
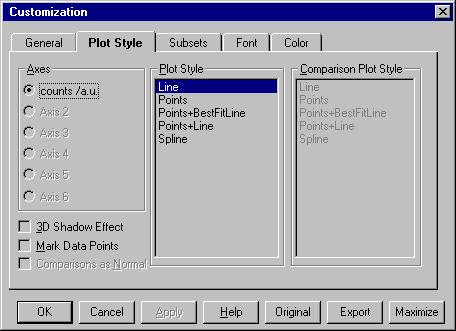
There are two interesting options here: Customization Dialog and
Export Dialog, both of them available also from the File menu. |
|
|
|
 |
|
|
|
.
By Customization Dialog (equivalent with File/Customize
Graph) we open the following menu. |
|
|
|
 |
|
|
|
The most important command here is Subsets
which is described later.
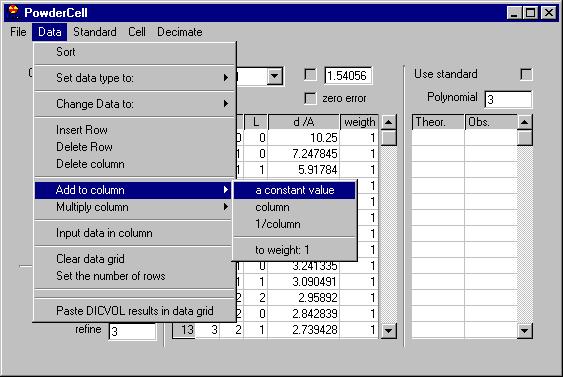
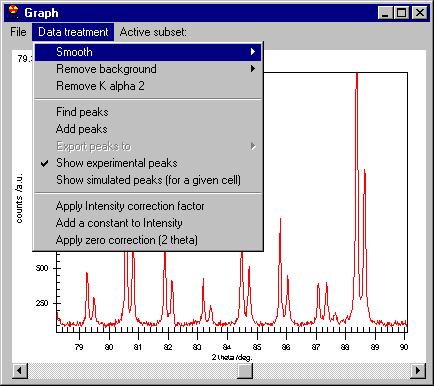
The second menu is Data treatment which allows:
-smooth by Savitzky Golay, Adjacent averaging or a Moving Window algorithm;
-remove background (automatic or manual),
-remove K alpha 2,
-Find peaks,
-Add peaks (not shown here but removing some of the peaks can be done easily,
just click on the marker)
-Export peaks (when you have already determined them)
-Show experimental peaks
-Show simulated peaks
-Intensity correction
-zero correction |
|
|
|
 |
|
|
|
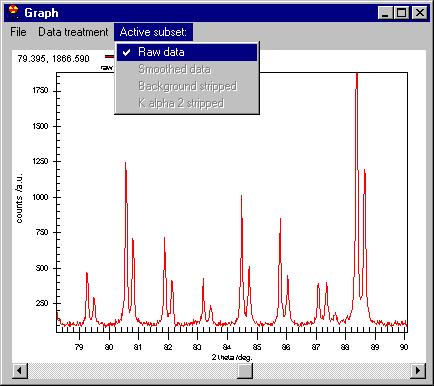
These commands are to be applied to the Active
subsets, i.e. |
|
|
|
 |
|
|
|
If Active subsets/Raw data menu is checked
then smoothing will affect the raw data (Note: you will not loose your original
data, all these are applied only in memory). You can smooth the already smoothed
data ("massaging" the data if you wish to do so) by checking Active
subsets/Smoothed Data.
Let's apply smoothing by Data treatment/Smooth/Savitzky Golay method
(a parabola through 7 points), after several seconds we get the following
screen: |
|
|
|
 |
|
|
|
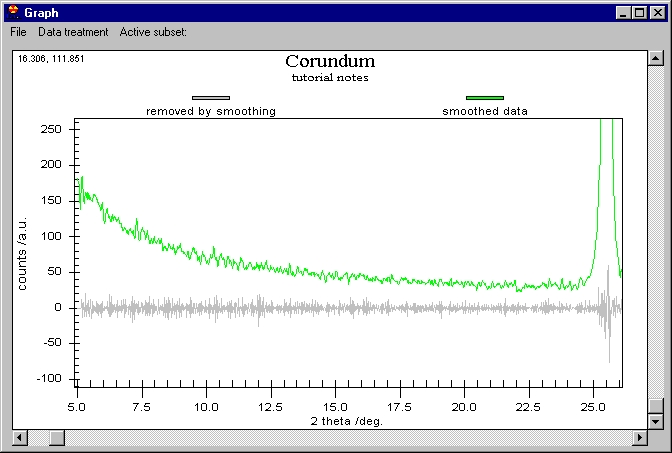
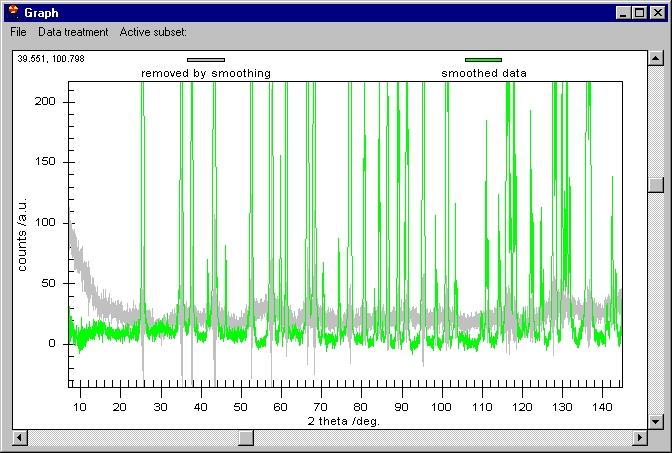
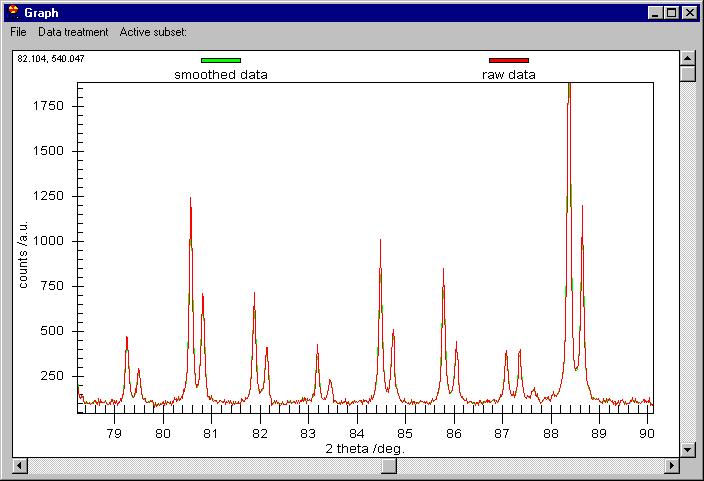
Although it seems that nothing was done, the
smoothed data are behind the raw data. You can change this or you can see
the difference between the smoothed data and the raw. Note the vertical scroll
bar here, it is not for zoom but for Scrolling between subsets. If we click
on the vertical scroll bar we obtain this: |
|
|
|
 |
|
|
|
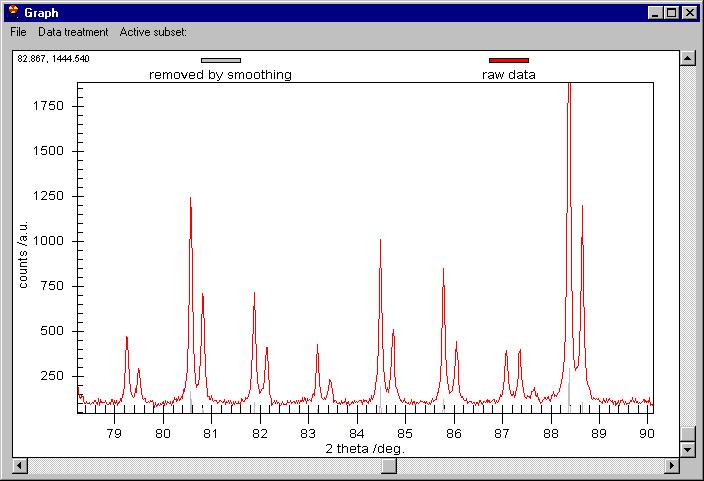
The Raw data are kept on the screen but smoothed
data have been changed to Removed by Smoothing. The Customization Dialog
controls which graph is shown.
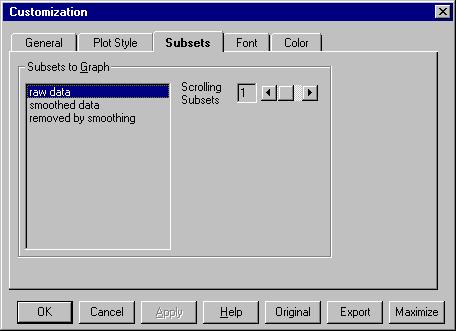
If we go now to Customization Dialog we can see this: |
|
|
|
 |
|
|
Raw data will be kept on the screen and we can
scroll 1 subset. If you select all the subsets or choose Scrolling
subsets equal with 2 you will see all the data sets.
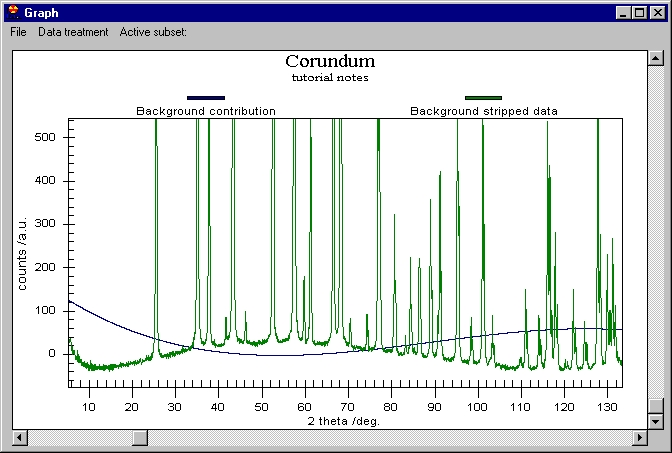
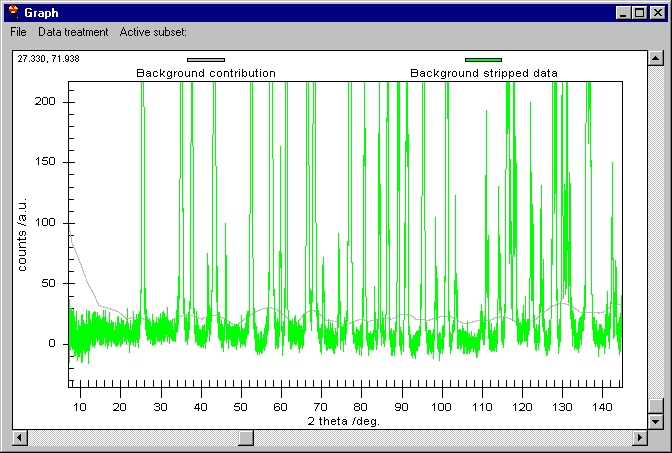
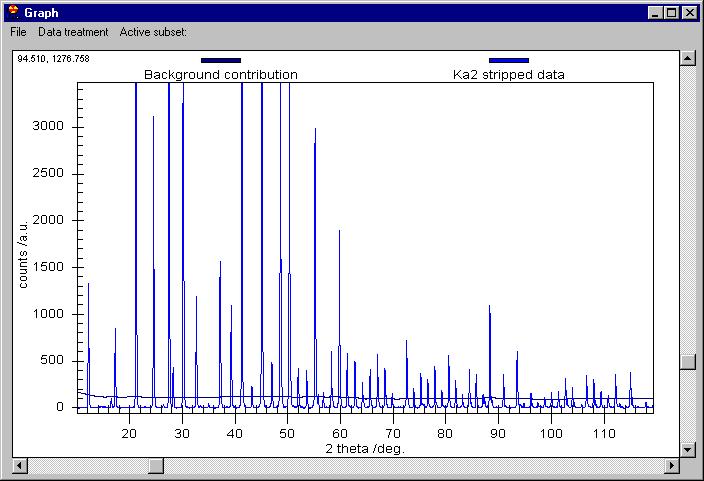
We can now remove the Background (note that Background removal applies to
the data which are selected in the Active subset menu) and then K alpha 2
to obtain this: |
|
|
|
 |
|
|
|
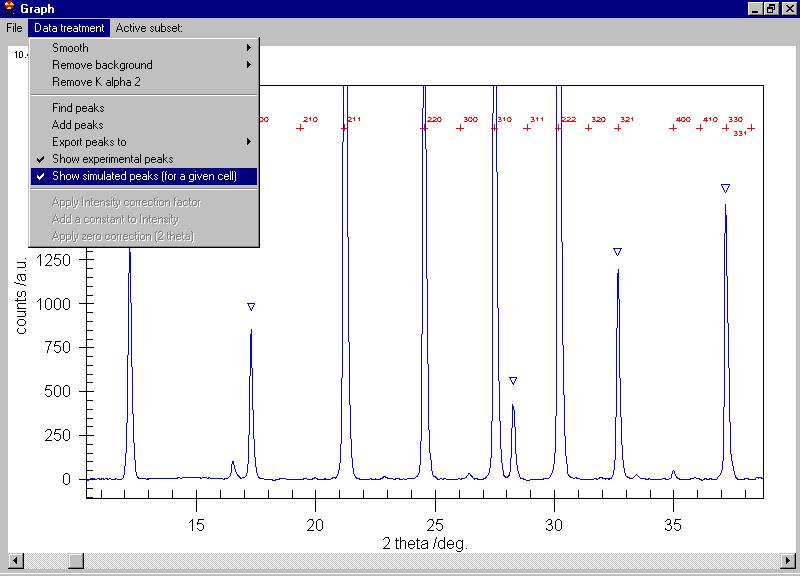
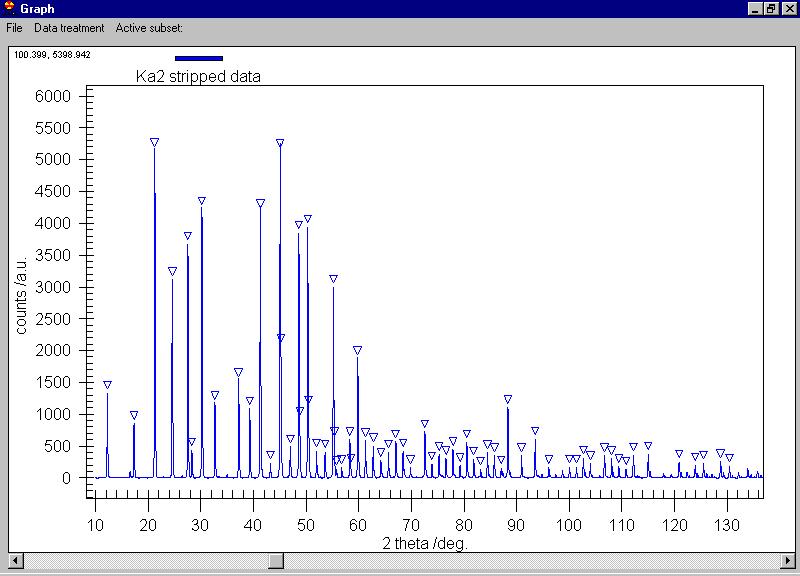
We can search for peaks and the peaks are marked
by triangles (some extra peaks appeared, you can delete them by clicking
on the triangle) |
|
|
|
 |
|
|
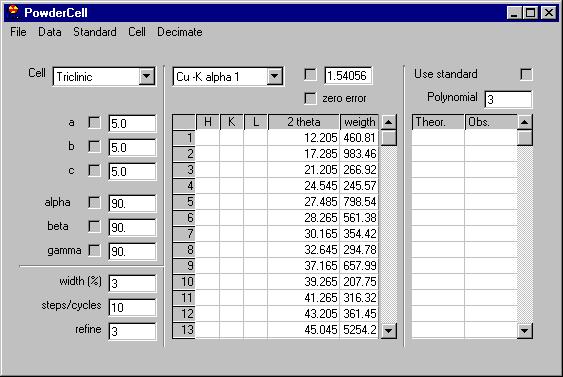
Removing several of the peaks which are obvious
due to noise (see the Rachinger method discussion in the book of Klug and
Alexander) we can send them all to the data grid, select the Export
peaks command: |
|
|
|
 |
|