The Rietbasic Script
Create a Rietbasic script to provide the automatic control (a PDF file is provided
with the Rietica distribution giving the list of Rietbasic options). As Rietbasic
is as a pseudo-programming language, it can be easier just to reverse-engineer and
modify an existing Rietbasic script.
Thus, modify the following Rietbasic macro script to match the name and number of
your data files (the script it is included in the zip file). You would change
the following within the script:
filestem = 'dat_'
startnum = 10
endnum = 20
The example data provided performs mass Le Bail fitting to get unit-cell
constants. If you were doing normal structure refinement, get rid of
the following line (which copies over the HKL file with the Le Bail intensities:
CopyFile(filestem+filenumber+'.hkl',filestem+filenumberx+'.hkl')
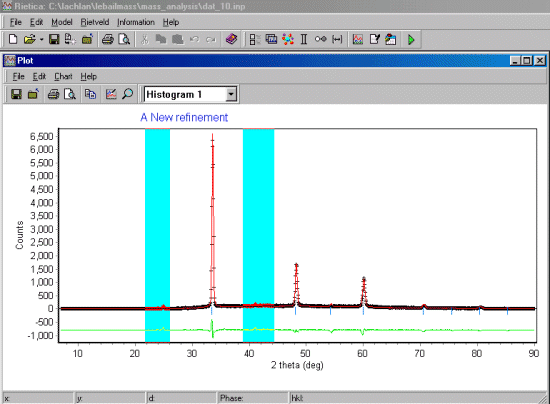
One limitation with this present script is there is no plotting of the data. You should
thus note how the fit parameter is varying and also look at the Rietveld plots after
to visually make sure everything has gone as intended.
Put the rietbasic RIB file you edited in the same directory where your data exists.
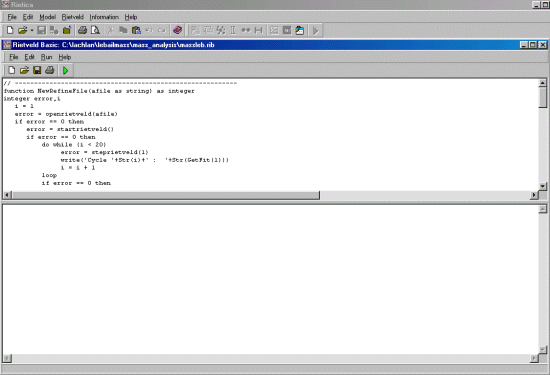
// ----------------------------------------------------------
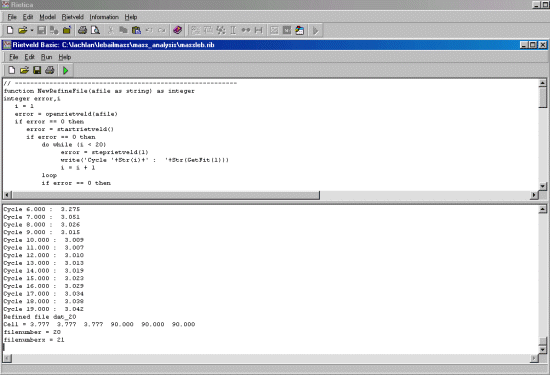
function NewRefineFile(afile as string) as integer
integer error,i
i = 1
error = openrietveld(afile)
if error == 0 then
error = startrietveld()
if error == 0 then
do while (i < 20)
error = steprietveld(1)
write('Cycle '+Str(i)+' : '+Str(GetFit(1)))
i = i + 1
loop
if error == 0 then
error = endrietveld()
// update() is undocumented call to update the inp file with the latest information
// Lachlan - be careful with this as potentially can corrupt starting input file
// if refinement goes beserk. Maintain a copy of you starting input file.
error = update()
end if
end if
end if
NewRefineFile = error
end function
// ---------------------------------------------------------
string workdir, filestem, filenumber,filenumberx
integer i,j,k,error,startnum, endnum
//------ run specific info -------------------
// "REM_this_out unless you want to" WorkDir = 'D:\rietica_analysis\dummy\'
filestem = 'dat_'
startnum = 10
endnum = 20
//--------------------------------------------
//write('')
j = 1
for i = startnum to endnum
filenumber = Str(i)
if i > 9 then
j = 2
elseif i > 99
j = 3
end if
filenumber = StrCopy(filenumber,1,j)
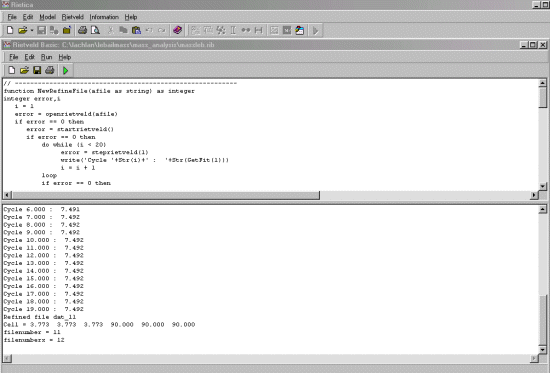
Write('Refining file '+workdir+filestem+filenumber)
error = NewRefineFile(workdir+filestem+filenumber)
if error == 0 then
write('Refined file '+workdir+filestem+filenumber)
write('Cell = '+Str(GetParameter(1, 1, 0, 6))+ ' '+Str(GetParameter(1, 1, 0, 7))+ '
'+Str(GetParameter(1, 1, 0, 8))+ ' '+Str(GetParameter(1, 1, 0, 9))+ '
'+Str(GetParameter(1, 1, 0, 10))+ ' '+Str(GetParameter(1, 1, 0, 11)))
k= i + 1
filenumberx = Str(k)
filenumberx = StrCopy(filenumberx,1,j)
Write('filenumber = '+filenumber)
Write('filenumberx = '+filenumberx)
CopyFile(filestem+filenumber+'.inp',filestem+filenumberx+'.inp')
CopyFile(filestem+filenumber+'.hkl',filestem+filenumberx+'.hkl')
else
write(GetError())
end if
next i