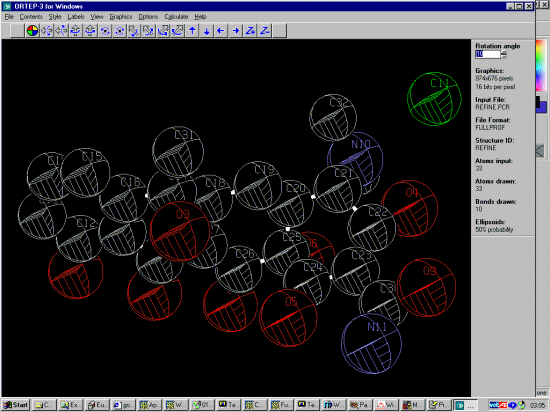
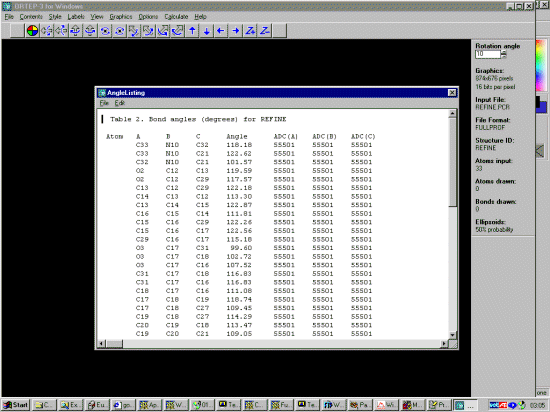
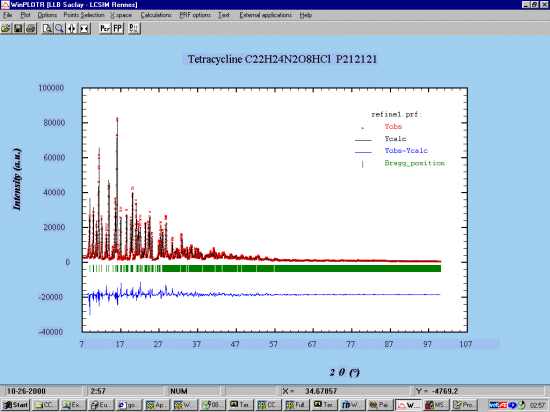
GSAS GUI WINORTEP by Louis Farrugia to examine the Structure and resulting bond lengths and angles during the refinement
Using Ortep-3 you can quickly load the Fullprof *.pcr file and check that the bond length and angle restraints are doing what you think they should be doing (using the Calculate menu option). You can also keep a graphical eye on the thermals.
- Ortep-3 - http://www.chem.gla.ac.uk/~louis/software/ortep3/ | [CCP14 UK Mirror | CCP14 Australian Mirror | CCP14 Canadian Mirror]
In the following case, thermals are quite large, possibly due to poorly defined manual background. Switching to a refined background might be a better thing to do here.