- To interactively visualise the map in 2D, run
Graphics, Forplot.
- Forplot has a wide range of functionality of which only a small subset
will be covered here.
Height of section above center is .000 A
The map size - center to edge is 5.00 A
The map grid interval is .300 A
Enter FORPLOT command (<?>,A,C,D,G,F,H,I,L,M,N,P,Q,R,S,T,V) >
FORPLOT commands:
<?> - Type this help listing
A - Define map center and orientation by entering 3 or 4 atom seq. numbers
C c - Set map center
D - Set atom labeling limit
F - Read a different Fourier map
G g - Set grid interval "g" in A (default = .3)
H h - Set height "h" of section above center in A
I v - Select contour interval in rho "v"
L - List current settings
M v - Select minimum rho value "v" (default = 0.0)
N n - Select number of contours "n" and assign their values
P - Plot map
Q - Quit FORPLOT
R a r - Enter axis (x,y,or z) and angle for rotation of current drawing
S s - Set map size - center to edge (default = 5.0 A)
T x y z - Display rho at "x,y,z"
V u v - Set map orientation vectors "u" and "v" (Enter 6 values)
Map horizontal is u and normal is uxv
Enter FORPLOT command (<?>,A,C,D,G,F,H,I,L,M,N,P,Q,R,S,T,V) >
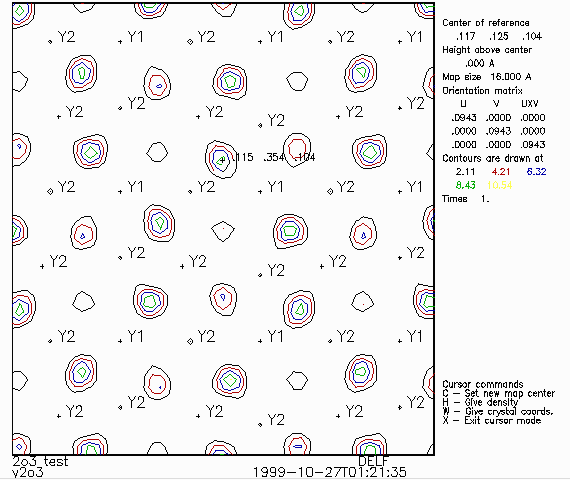
- For instance, to see where existing atoms are projected, you can use
D Set atom labeling limit to have atoms labeled a user defined distance from the slice
being viewed. (e.g., 2 Angstrom).
- H (Set height "h" of section above center in A) to match the above found
oxygen atom so you can visually look at the peak. In the following graphics window, you can
use the mouse and keyboard to graphically center the map on a desired area and obtain
co-ordinates.
|
