These are the principal features required in modern data treatment where an environment parameter (temperature etc.) is scanned, and the set of data can be treated together. The second case is highly relevant for neutron scattering data where selective deuteration can be used to emphasise features of otherwise similar samples.
A technical description is available to help programmers maintain the fitting suite.
The User's View of Clickfit
There are two versions of the program, Clickfit for treating one or sequences of individual datasets. Clickfit_m is adapted to treat the case of controlling simultaneous fitting of several datasets, where more selective control of fitting parameters is required.
Often fitting programs are written with several similar variants, or certain control parameters can be predefined for specific tasks. On launching Clickfit the first frame contains the editable list of program variants available. The most recently used program is highlighted.
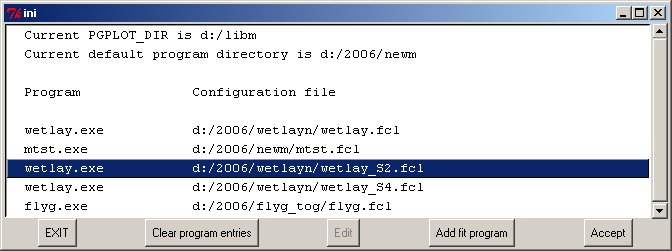
The programs use a number of common components; to simplify relocating onto other computers the fields here contain the important placement of the graphics package PGPLOT and the current default program directory. The latter would normally contain the executable programs and their clickfit configuration files and variants, and thus make it simple to re-install the set on another computer with a different directory structure. The SANS program package includes rfit.fcl, sasfit4.fcl, polymer.fcl, hs2.fcl, hs2m.fcl etc. Many fitting programs on the Reflectometry Site share descriptive help files (.hhh) for input etc. Once selected these options may be edited. Clicking on ACCEPT starts the selected program, passing to an optional PRELUDE page.
The Prelude
To allow programmers a wide choice of additional options before entering the principal part of the fitting program a terminal program will often pass through a dialogue phase to enter special parameters.common/guiuse/ignum,insta,iosta,irsta common /comin/ bufin character bufin*150 : call prelude(pnam) if(ignum.le.0) .....use terminal for flayer,datup,rwt,rq4,rres ....otherwise read from bufin read(cbufin,*) flayer,datup,rwt,rq4,rresFor Clickfit there is a configurable prelude section where values can be entered for later use. Here the options for wetlay are shown:
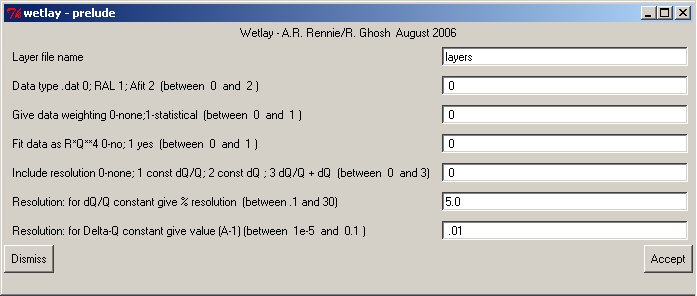
After any edition pass to the next step with the ACCEPT button.
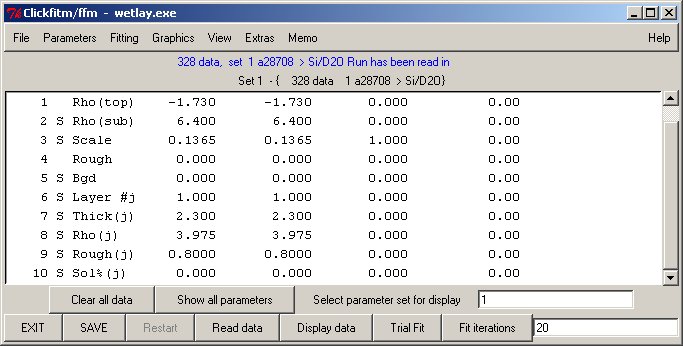
Functions offered by Clickfit
Essentially the same GUI interface routines can control either single data fits (using library fitfun v8.0) or simultaneous fitting of several sets (fitfun_m v8.1) This section offers most of the original features of the fitfun command line.
Menu options
File
Save parameters into a *.ffn or *.ffm file, used on restart.
Append to list file outputs current fit information to listfile
Read in brings up data input panel
Exit terminates program
Parameters
edit parameter edit parameter and step
Link steps parameters grouped to be varied with the same step
Copy parameter set* copies parameters from one dataset for another
Fitting
Limit x Ranges Restrict fitting to three x ranges (max)
Step size The fractional step multiplied by the parameter
step size for numerical derivatives
Graphics
Set scales sets scale limits for x and y
Append PostScript plot add a plot of the current fit to file
Cursor toggle cursor on/off in next display
Title Add title to next plots
Write plot datafile Write out fits and data as ascii file
View
View log Displays current log file
View sets* lists current sets
Extras
Special Function (J command - often a display/output
option
Command Access to basic fitfun command line
Debug Toggles on and off - shows comms
Memo up to 4 manual commands can be stored/recalled
Help Help file (or other file if configured)
Modify parameters Double click on parameter line
Buttons Clear data* resets all sets Show all parameters* displays parameters for all sets Select set for display* give set number for current display Fit Sequence+ fit a sequence of data using input control file Make Sequence+ will filter a sequence of filenames to treat SAVE SAVE current parameter (reread at next start) Restart when configured - restart prgram Display Data display current data (with/without cursor) Trial Fit Show current model calculation Fit Iterations iterate the indicated (max) number of times EXIT EXIT program (please AVOID terminating window) + Clickfit * Clickfit_mData treatment proceeds through the stages of READ/Trial/Iterate../(SAVE)/EXIT
Sequences
Before a sequence can be treated an initial dataset must be fitted to set the initial parameters to approximate values for the sequence. The information for each member of the sequence is read from a control file; a simple example follows:
Fileseq.fsq... gtst sequence title fitseq.out output filename T Reuse first trial (T) or update (U)parameters for next set 50 Number of iterations r.spc 3.5 input data for each fit s.spc 4.2 : t.spc 4.9 :When (typically) only a filename is used for each set of data to be treated the Make sequence button will bring up a window to create a control file automatically, which is then used to treat the (filtered) filenames therein:
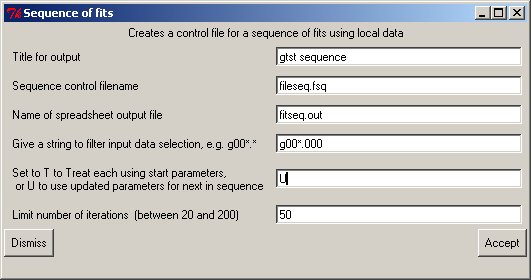
The filter allows an initial set of files to be chosen; this leads to the next window to allow a selection and order to be completed with the panel allowing single/multiple selects etc., and the fits performed.
Installation and Use
The program scripts may be found at:clickfit.zip
Clickfit requires the wish/tcl/tk package and graphics, using pgplot, also must be installed. When clickfit runs for the first time it will need to informed of the default directories for programs and work
PC and Windows
The program is started as a batch file (.bat) It is assumed that the program is run using the prop package and this contains all the prerequisites. The two files clickfit.bat and clickfit_m.bat are simply copied to the project program directory and (if necessary) added to the program selector.
Unix/Linux/OSX
The plotting environment variable PGPLOT_DIR should be set to find the PGPLOT xserver program and fonts. The clickfit.bat and clickfit_m.bat files may require editing on the first line to use the local version of wish, alternatively they may be started with the command
% wish clickfit.bat
Acknowledgements
The routines have been tested with the suite of programs for neutron reflectivity data analysis programs and the encouragement of the author, Professor A. Rennie who has made them available for use at the ILL, is gratefully acknowledged here.