Version 3.5
R. Ghosh, May 2004
The detector efficiency is usually measured using a uniform scatterer. Typically a 1mm water sample is used with the detector at a few degrees offset from the direct beam. By dividing the measured spectrum by the water spectrum the relative cell by cell efficiency and the solid angle subtended by each cell are taken into account. If no normalising run is give then the efficiency is taken as unity everywhere.
The program red16 can treat one or a sequence of frames (which may have consecutive or uniformly spaced run numbers).
The data in each frame are added into a resultant spectrum, taking account of any changes in the detector angle. The multi-detector data can be added using an equatorial projection (which simply corresponds to adding the data in columns of y), or azimuthally, which takes into account the scattering on cones centred at the sample. Individual cell contents are allocated to adjacent uniform mesh values using fractional linear interpolation from the cell position.
Regrouping is made in equi-angle steps; by default the x-pixel width is converted to the angular step width.
Parameters and efficiency data which have been changed are written to disk and are re-used the next time the program is run.
The output listing includes the statistical error in the intensity per cell, and also the weighted deviation for the contributing cells. For a uniform distribution these are similar; large differences indicate sample ordering giving rise to anisotropy.
R run1 [run2] [da] [dr] Regroup dataRegroups data for runs run1 to run2, with a step of dr to allow for repetitive non-contiguous sequences. The angular step, da degrees, overwrites the default angular step derived from the x-pixel width.
The present version of the program uses the monitor of the first run of a sequence to re-ratio subsequent scan components. Thus if measurements are made using constant time intervals a small correction is made. A table of count rates and the ratios of components flux to that of the first scan is shown. If these show strong deviations from constancy then this should be further investigated. Normally the reactor flux is constant to better than 1%.
Including sets of runs
R [I] [T] run1 [run2] [da] [dr]Successive use of RI commands add the runs or sequences to a list for treatment together. If the letter T is included then this terminates the list and treatment commences.
If, after starting a list an error is found, then the command R alone will reset the number of runs to zero and no treatment will be performed.
L run1 run2 lset [da] >This will reduce data for linked sequences of runs length lset starting at run1 and saving the reduced data as SANS data files. The next sequence is a set of the same length starting at run1+lset-1, etc., if this is less than or equal to run2.
N run ext low high Normalisation dataTakes the data from run/ext and assumes this is a flat spectrum to produce a file of efficiency dividing factors with an average value of unity. If the limit levels are non-zero, the normalised factors searched, only including pixels within values, and the normalisation corrections are recalculated for these remaining data. These dividers are stored. If a zero run number is given then unit efficiency is assigned to all cells.
Using other programs from the SANS suite, e.g. anpoly, effdef and rmask it is possible to perform more complex masking than the simple window included in the program.
D r1 r2 r3 r4 .. Raw runs from a nominally isotropic sampleThe raw data are summed and a normalisation factors for the detector efficiencies are calculated and stored for subsequent use. Up to ten runs may be included.
B x0 y0 g0 sd wav Beam centre, s-d distance, wavelengthThe incident beam characteristics are set with this command, namely the beam centre, x0 and y0 (in cells) when the detector angle is at angle g0, the sample-detector distance (in metres), and the wavelength in Angstroms. If the data contain a significantly different wavelength a message is printed out, but this wavelength, if set, is used instead.
M maskfile Reads in a maskfileA maskfile constructed with standard SANS programs rmask, grund etc can be read in and used to designate cells to be eliminated from regrouping calculations. A blank filename eliminates the mask option. Note: the name and extension must be given; both are case-sensitive.
A [A] or [E] Averaging mode for regrouping - azimuthal averaging
or equatorial projection
This command sets the type of regrouping procedure to be used.
G [H] [V] or [S] Detector Geometry Horizontal or Vertical
or Square for the new 16K detector
This command sets the appropriate cell dimensions and orientation.
F filename Ascii file of Angle/Counts/Error/PixelsThe regrouped data from the R command are written into a file "filename". Three header information lines are followed by the number of points and then four columns of angle, counts, the standard error in the counts, and the number of contributing pixels.
S save data in ILL-SANS format Q/Counts/ErrorThe regrouped data are stored in standard SANS format and may be treated with programs from this suite.
P [H] Plot data as Angle/Counts/ErrorThe regrouped data are plotted within the limits set by the X and Y commands. If the letter H is included then the data are written out for printing (typically as a PostScript file, though this depends on the setting of the environment variable PGPLOT_ILL_DEV_2)
Q [H] Plot data as Q/Counts/ErrorThe regrouped data are plotted within the limits set by the X and Y commands. If the letter H is included then the data are written out for printing (typically as a PostScript file).
X [min] [max] shows or sets the X scale limits for plots Y [min] [max] shows or sets the Y scale limits for plots C toggles cursor off or on in the next plotClicking in the graphics window, or pointing and typing spaces marks the coordinates of the cursor on the graph. These markers are reproduced on hardcopy if the next plotting command includes an H. The end of cursor input is signalled by typing RETURN while the pointer is in the graphics window.
W [R] [x1 x2 y1 y2] show or [Re]sets data window to be usedThe rectangle defined by the coordinates x1,x2,y1,y2 only is used for regrouping. This is a simple way of eliminating the outside regions, if these are considered to contain unreliable data. The first and last rows and columns are always excluded from all regrouping.
I r1 [low high] [H] [R] displays detector imageThis is shown within the low and high limits if set. The run number is incremented, so a second I command would show the next run. Normally the efficiency corrected and masked data are shown. If R is also given in the command then the raw data are shown. Adding H to the command produces a hardcopy file. Giving low a negative value resets automatic scaling within the data limits.
V shows parameters set for current treatment
Z resets all options to default values
This shows sequence being treated, missing one frame, and then plotted.
is1 11% dirrun /usr/illdata/data/d16/d16_5
Data in directory /usr/illdata/data/d16/d16_5
numors 050000 to 55101
5101 data files are stored here
is1 12% setenv SAS_DATA_PATH /usr/illdata/data/d16/d16_5/
is1 13% red16
red16 - version 3.5 May 2004 (R. E. Ghosh)
reduces D16 scan data
Options:
R run1 run2 da dr Reduce run1 to run2 angle-step run-step
R[I][T] r1 r2 da dr Include runs r1 to r2 etc for treatment
if [T] is present the treatment commences
if r1 is <=0 the list is cancelled
L r1 r2 lset da Linked sets length lset, reduce to SAS
output files all groups starting r1 to r2
N run ext low high Normalisation input for detector efficiency
if set only use values between low and high
D r1 r2 r3 r4 .. Detector raw data for efficiency corrs.
up to 8 runs may be read in
B x0 y0 g0 sd wav Beam centre(set when non-zero values given)
x0,y0 cell#, g0 offset, sd dist.(m), wav A
M maskfile Mask data with maskfile
A [A] or [E] Average azimuthally or equatorial proj.
G [H],[V] or [S] Geometry of detector, horiz/vertical/square
S save data in SANS format
F filename File as ascii data (angle)
P [H] Plot results (angle) [Hardcopy]
Q [H] Plot results (Q) [Hardcopy]
X [min] [max] shows or sets x scale limits
Y [min] [max] shows or sets y scale limits
C toggles cursor in next plot
W [R][x1 x2 y1 y2] show or (Re)sets window on data
I r1 [low high] image of run r1 shown in limits
give I alone to show next run
add H - Hardcopy, R - show raw data
V show values used in treatment
Z reset all values to defaults
H HELP (this text)
EXIT end program
Norm. Runs 54872 58473 58474 58457
Deviation of normalisation data 11.9
Number of pixels eliminated 508
Thresholds between 0.00 and 0.100E+11
Mask: ** No Mask **
Title:
Number of pixels outside data window 0
Beam centre cells X0 63.30 Y0 63.50
Detector offset zer0-angle G0 0.00
Sample-detector distance 1.000m
Wavelength 4.5400A
Azimuthal regrouping to be used
Square detector in use
Option R,L,N,D,B,P,M,A,G,Q,S,F,X,Y,W,I,V,Z HELP or EXIT :
red16> r i 55000 55004
There are now 5 runs ready for treatment
Give option R,L,N,D,B,P,M,A,G,Q,S,F,X,Y,W,I,V,Z HELP or EXIT :
red16> r it 55006 55008
There are now 8 runs ready for treatment
1 sap0.7 D2O 5
Sample-detector distance 1.000m
Wavelength 4.5400A
Run Angle Monitor Time Mon. rate Mon/Mon1
55000 30.00 327301. 100.00 3273. 1.00000 sap0.7 D2O 5
55001 32.03 328866. 100.00 3289. 1.00478 sap0.7 D2O 5
55002 34.00 328476. 100.00 3285. 1.00359 sap0.7 D2O 5
55003 36.00 328883. 100.00 3289. 1.00483 sap0.7 D2O 5
55004 37.99 329104. 100.00 3291. 1.00551 sap0.7 D2O 5
55006 42.01 327546. 100.00 3275. 1.00075 sap0.7 D2O 5
55007 44.00 329108. 100.00 3291. 1.00552 sap0.7 D2O 5
55008 46.01 328385. 100.00 3284. 1.00331 sap0.7 D2O 5
Angle Q Counts Error W-deviation Pixels
22.80 0.5472 2.51 1.16 0.85 2.1
22.92 0.5499 1.75 0.24 0.23 33.5
23.03 0.5526 1.78 0.19 0.18 54.4
23.15 0.5553 1.65 0.16 0.13 69.2
23.26 0.5580 1.83 0.15 0.15 81.3
23.38 0.5607 2.02 0.15 0.15 92.1
23.49 0.5635 1.95 0.14 0.14 102.0
23.61 0.5662 2.10 0.14 0.14 110.8
23.72 0.5689 2.06 0.13 0.14 119.2
:
40.68 0.9621 1.49 0.04 0.04 765.2
40.79 0.9647 1.54 0.04 0.04 765.1
40.91 0.9673 1.62 0.05 0.05 765.2
41.02 0.9699 1.65 0.05 0.05 765.2
41.14 0.9725 1.68 0.05 0.05 765.2
41.25 0.9751 1.63 0.05 0.05 729.7
41.37 0.9776 1.77 0.05 0.05 693.1
41.48 0.9802 1.85 0.05 0.05 670.3
41.60 0.9828 1.91 0.05 0.05 652.0
41.71 0.9854 1.83 0.05 0.05 638.2
41.83 0.9880 2.07 0.06 0.06 636.5
41.94 0.9906 2.22 0.06 0.06 636.6
42.05 0.9932 2.35 0.06 0.06 636.6
42.17 0.9958 2.61 0.06 0.07 636.5
42.28 0.9983 2.83 0.07 0.07 636.5
42.40 1.0009 3.17 0.07 0.07 636.6
42.51 1.0035 3.51 0.07 0.08 636.6
42.63 1.0061 3.81 0.08 0.08 636.6
42.74 1.0087 4.13 0.08 0.08 636.7
42.86 1.0112 4.21 0.08 0.08 636.9
42.97 1.0138 4.22 0.08 0.08 636.9
43.09 1.0164 4.02 0.08 0.08 636.9
43.20 1.0190 3.63 0.08 0.08 623.4
43.32 1.0215 3.28 0.08 0.08 575.8
43.43 1.0241 2.99 0.07 0.08 548.7
43.54 1.0267 2.64 0.07 0.07 527.9
43.66 1.0292 2.32 0.07 0.07 511.6
43.77 1.0318 2.15 0.06 0.07 508.4
43.89 1.0344 1.91 0.06 0.06 508.4
44.00 1.0369 1.77 0.06 0.06 508.5
44.12 1.0395 1.66 0.06 0.06 508.5
44.23 1.0421 1.53 0.05 0.06 508.5
44.35 1.0446 1.46 0.05 0.06 508.6
44.46 1.0472 1.47 0.05 0.06 508.8
44.58 1.0498 1.44 0.05 0.06 508.8
44.69 1.0523 1.36 0.05 0.05 508.8
44.80 1.0549 1.33 0.05 0.05 508.9
44.92 1.0574 1.26 0.05 0.05 509.0
45.03 1.0600 1.20 0.05 0.05 509.1
45.15 1.0626 1.23 0.05 0.05 507.5
:
52.48 1.2238 1.03 0.09 0.09 128.2
52.60 1.2263 1.19 0.10 0.10 128.2
52.71 1.2288 1.16 0.10 0.09 128.3
52.83 1.2313 1.19 0.10 0.09 128.3
52.94 1.2338 1.16 0.10 0.09 128.4
53.06 1.2362 0.93 0.09 0.08 128.4
53.17 1.2387 0.84 0.09 0.09 126.4
53.28 1.2412 1.18 0.15 0.14 71.7
53.40 1.2437 1.07 0.21 0.15 33.2
53.51 1.2461 0.85 0.41 0.36 7.1
Option R,L,N,D,B,P,M,A,G,Q,S,F,X,Y,W,I,V,Z HELP or EXIT :
red16>c
Option R,L,N,D,B,P,M,A,G,Q,S,F,X,Y,W,I,V,Z HELP or EXIT :
red16>p
LEFT MOUSE OR TYPE SPACE FOR X,Y OR RETURN TO CONTINUE
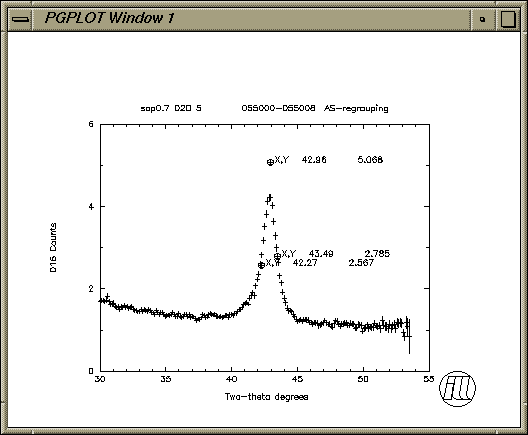
Option R,L,N,D,B,P,M,A,G,Q,S,F,X,Y,W,I,V,Z HELP or EXIT : red16>p h Option R,L,N,D,B,P,M,A,G,Q,S,F,X,Y,W,I,V,Z HELP or EXIT : red16>e Closing listing file d16r637.lis %PGPLOT, Closing file d16r638.ps is1 14% plo d16r638.ps request id is hp_cs-1191 (1 file(s))
SAS_INSTRUMENT set to d16
SAS_DATA_PATH set to directory containing raw data
NOTE: this should have a terminating / (Unix)
or \ (PC)
SAS_DECOMPRESS set to zcat if automatic data decompression is
to be invoked
Plotting control
PGPLOT_DIR set to /usr/ill/bin for font file etc.
PGPLOT_ILL_DEV_1 set to /XSERVE for X-window graphics, /NULL none
PGPLOT_ILL_DEV_2 set to /VCPS for PostScript plots
PGPLOT_ILL_PPAGE set to 2 for 2 pictures per page